AbstractThe bacterial community structure of polluted soil differentiates according to toxic pollutants. In this study, the acid pollution source was predicted by using characteristic bacterial community structures which were exposed to HCl, HF, HNO3, and H2SO4. In a soil column, after a simulated acid leak, Bacillus, Citrobacter, Rhodococcus, and Ralstonia sp. were found as acid-resistant bacteria and their relative abundance varied depending on the acid. The complex bacterial community was analyzed by using terminal restriction fragment length polymorphism (T-RFLP) of 16S rRNA gene. Using machine learning models including support vector machine (SVM), K-nearest neighbors (KNN), random forest (RF), and artificial neural network (ANN), the prediction accuracy for acidic pollutants was 72%, 72%, 76%, and 88%, respectively. With data augmentation based on T-RFLP, the accuracy of the ANN model for predicting acidic pollutants improved to 98%. This research provides valuable insights into the potential use of bacterial community structures and machine learning models for the rapid and accurate identification of acid pollution sources in soil.
Graphical Abstract
1. IntroductionSoil is a vital part of our planet, comprising 25% air, 25% water, and the remaining 50% being a mix of organic and inorganic materials [1]. This soil offers us essential services, from food and fuel to building materials, supporting both our health and ecosystems [2]. However, mining and manufacturing industries are rising as major causes of soil pollution, which is becoming a big issue [2, 3]. Contaminated soils pose risks to both ecology and human health, intensifying the need for effective methods to identify and track such pollution [4–7]. Moreover, the leakage of chemicals can change the soil’s pH and other important features, highlighting the need for studies on these effects and urgent soil protection efforts [8–10].
Bacterial communities within soil are crucial for maintaining its health and functionality [11–13]. These changes in the soil bacterial community can be seen as sensitive indicators of environmental changes and contamination events [14, 15]. Specifically, Proteobacteria play key roles in environmental processes like nitrogen fixation [12], while Firmicutes are known to be active even in certain acidic conditions [14, 16]. Analyzing the structure of these bacterial communities can serve as an essential tool for understanding and addressing soil contamination. Terminal restriction fragment length polymorphism (T-RFLP) stands out as a cost-effective technique useful for assessing such bacterial diversity, offering a concise depiction of bacterial classifications [17, 18]. Therefore, a detailed analysis of bacterial communities plays a significant role in evaluating soil’s environmental health and contamination status.
Soil pollution, particularly acid contamination, arises from various sources and negatively affects the environment. In a previous report, numerous factors, including industrial accidents, have been pinpointed as leading causes of soil pollution in the industrial sector [2]. Chemical spills, fires, and explosions can result in major disasters for residents and the environment, causing financial losses, raw material shortages, and severe disruptions across various industries [19–21]. HCl can easily induce soil and groundwater acidification [22, 23] while HNO3 reduces soil’s buffering capacity [24]. In addition, H2SO4 and HF cause soil expansion and rapidly dissolve plant silica, respectively [25]. Such acid leaks lead to environmental problems like a decrease in soil pH, resulting in shifts in soil bacterial communities, reduced crop yields, and respiratory diseases [4, 9, 26–28]. Considering these multifaceted causes and impacts, it is essential to accurately identify acid pollution sources.
Soil contamination is becoming increasingly complex, necessitating the use of modern technologies. Machine learning models, such as artificial neural networks (ANN), random forest (RF), support vector machines (SVM), and K-nearest neighbors (KNN), can be utilized in predicting acidic pollution sources [29–32]. Inspired by the structure of the human brain, ANN are capable of processing complex data information arising in genetics, such as the 16SrRNA gene of bacteria, enabling the prediction of relationships with acidic contaminants [29, 33, 34]. However, to enhance the accuracy of these predictions, a sufficient amount of data is required for training machine learning models. The introduction of generative adversarial networks (GAN) for data augmentation provides a solution to this challenge [35, 36]. In comparison to conventional machine learning, GAN can greatly refine the accuracy of identification of soil contaminants. In this study, it was hypothesized that machine learning models combined with tools like T-RFLP enhance the performance of forensics by generating diverse biological indicators of terminal restriction fragments (T-RFs) [17, 37–39]. Harnessing a blend of these advanced techniques and data methodologies holds great promise in addressing the challenges of soil pollution.
This study aims to develop a forensic tool for acidic pollution through T-RFLP analysis and machine learning. After introducing tap water, HCl, HF, HNO3, and H2SO4 to healthy soil, we investigated the temporal changes in bacterial communities using both T-RFLP and next-generation sequencing (NGS). The data obtained from T-RFLP was augmented using GAN. Then, using trained machine learning models, including SVM, KNN, RF, and ANN, we classified the samples based on the specific acidic pollutants. This novel forensic approach offers precise predictions and insights into the effects of various acidic contaminants on bacterial communities.
2. Materials and Methods2.1. Simulated Acidic Pollutant LeakageTo simulate an acidic contaminant leak, 10 soil columns with a diameter of 5 cm and a length of 21 cm were filled with 550 g of soil (from 35°14′38.4″N 129°03′23.4″E). Subsequently, tap water (pH 7.01±0.1, conductivity 98±10 us/cm), 1.0 N HCl (Samcheon Chemical Co., Korea), HF (Duksan Pure Chemical Co., Korea), HNO3 (Samcheon Chemical Co., Korea), and H2SO4 (Junsei, Japan) were added to the five autoclaved soil columns in volumes equal to the soil. The same process was also performed on the five non-autoclaved soil columns with the same acidic contaminants. Leakage was conducted on days 1 and 3. From day 5 to 10, rain simulation was performed using an amount of tap water equivalent to the soil volume. Starting from day 10, this rain simulation was repeated every 5 days using tap water of the same specifications. In this manner, a total of 34 days were spent conducting leakage and rain simulations.
2.2. SamplingFrom the soil column subjected to tap water and four types of acidic contaminant leakage, soil samples were collected on days 1 and 3 post-leakage, during the rain simulations from days 5 to 10, and on days 16, 22, and 28. Additional samples were also taken on days 19, 25, 31, and 34 when no treatments were applied. On days 1, 3, and the final day 34, samples were specifically collected from the top, middle, and bottom sections of the soil column, totaling 100 soil samples (Table S1).
2.3. pH AnalysisEach 2.5 g soil sample was mixed with 5 mL distilled water, agitated at 150 rpm, 25°C for 30 minutes, and its pH was measured using a pH probe (AB15+ Model, Thermo-Fisher Scientific, USA).
2.4. DNA ExtractionSoil samples for DNA extraction were collected from the soil column, amounting to 500 mg, and were stored at 20°C until DNA extraction. A total of 100 samples were collected, and sample codes are provided in Table S1. Genomic DNA was extracted using the SPINeasy DNA Kit for Feces/Soil (MP Biomedical, USA), and further purified with the DNeasy Power clean Pro Cleanup Kit (Qiagen, USA). The concentration of the extracted DNA was analyzed using the QubitTM 4 Fluorometer (Invitrogen, USA).
2.5. Next-Generation SequencingFor the analysis of soil bacterial communities, Bacterial 16S rRNA gene amplicons (V3 and V4 regions) were sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA, United States) by Macrogen Inc (Seoul, South of Korea). Post-sequencing, samples were categorized using index sequences, and paired-end FASTQ files were generated. Sequencing adapters and target gene region primers were removed with Cutadapt (v3.2). Amplicon sequencing was error-corrected using the DADA2 (v1.18.0) package in R (v4.0.3). Reads exceeding 2 expected errors were discarded, and sequences were truncated to 250 bp for Read1 and 200 bp for Read2. After establishing a batch-specific error model, noise was removed from samples. Corrected paired-end sequences were merged and chimeric sequences were removed using DADA2’s Consensus method to form ASVs. For microbial community comparisons, normalization was performed using QIIME (v1.9) based on the sample with the fewest reads. Each ASV was matched against the NCBI 16S Microbial DB using BLAST+(v2.9.0) to assign taxonomy, but assignments were skipped if query coverage or matched region identity was below 85%.
2.6. Terminal Restriction Fragment Length PolymorphismThe 16S rRNA gene was amplified for T-RFLP analysis using primers 27F and 518R [40, 41], labeled with FAM and HEX fluorophores. The PCR mixture of 50 μL included DreamTaq DNA Polymerases (Thermo Fisher, USA), primers, DNA template, and deionized water. Amplification occurred in a SimpliAmp Thermal Cycler (Thermo Fisher, USA) with specific cycling parameters: initial 3 min at 93°C, followed by 30 cycles of 30 s each at 93°C, 60 °C, and 72°C, concluding with a 10 min cycle at 72°C. After amplification, the amplicon was purified using QIAquick PCR Purification and digested with endonuclease BsuRI (Thermo Fisher, USA) at 37°C for 3 hours. T-RF scanning was performed by SolGent (South Korea) with data processed using the Peak Scanner (Thermo Fisher, USA). T-RFs were sorted, and their relative abundance was determined using the “readr”, “dplyr”, and “tidyverse” packages in R.
2.7. Indicator Species AnalysisThe T-RF data obtained through T-RFLP analysis was utilized for the assessment of indicator species specific to certain acid pollutants. The T-RFs and their relative abundance from each soil sample were organized into a data frame format. After standardizing this data frame, it was analyzed using the ‘Indicspecies’ package in R. Based on the analysis, data on indicator species for each acid pollutant and other species were separately extracted.
2.8. Data AugmentationData augmentation was carried out using Python. The numpy and pandas libraries were employed for data processing, and Keras was utilized to establish a generative adversarial network (GAN) model. The data frame was organized such that the first column represented samples, the second column denoted acidic pollutants, and the columns from the third onward contained data. Additionally, standardization was performed on this data. For the model, the generator was designed with an initial input of 100 nodes, followed by layers with 256, 512, and 1024 nodes, respectively. The final layer contained nodes equivalent to the number of features in the original data, employing a ‘linear’ activation function. The discriminator was constructed with layers housing 1024, 512, and 256 nodes sequentially, with its final layer utilizing a ‘sigmoid’ activation function. During model training, the Adam optimization algorithm was used. The learning rate, set at 0.0002, controlled how swiftly the model learned, ensuring stable training. Additionally, a beta1 value of 0.5 was set, determining the extent to which past gradient information was reflected. The configured model underwent training for a total of 1000 iterations (epochs). Using the trained generator, 20 new sample data points were generated for each acidic pollutant.
2.9. Machine Learning for Pollutant PredictionVarious machine learning algorithms were experimented with to predict acidic pollutants. The raw data was loaded using the pandas package and preprocessed with numpy. After removing missing and outlier values, the data was standardized and split into training and testing sets at an 80:20 ratio. The support vector machine (SVM) was trained using sklearn with a linear kernel and C=1. For the K-nearest neighbors (KNN), the number of neighbors was set to 3. The random forest (RF) was configured with 100 trees, and the artificial neural network (ANN) was designed using tensor-flow with two dense layers (128 and 64 neurons) and a softmax output layer. The performance of each model was evaluated using accuracy, classification reports, and confusion matrices. For visualization, 2D PCA, neural network diagrams, and confusion matrices were used.
3. Results & Discussion3.1. Changes in Soil pH Due to Acidic ContaminantsTo monitor the soil pH over time following the acidic substance leak, tap water (W-column), HCl (C-column), HF (F-column), HNO3 (N-column), and H2SO4 (S-column) were leaked into both the five non-sterilized soil-filled columns and the five sterilized soil-filled columns on days 1 and 3. Subsequently, using tap water, periodic rainfall was simulated for 34 days, and the top layer of soil from the columns was periodically collected to measure the pH.
As a result, after the acidic contaminant leak, a rapid decrease in pH was observed in both the five sterilized and the five non-sterilized soil samples. In both conditions, the soil showed an increasing pH trend for about 10 days, after which the pH in the soils with the leaked acidic contaminants stabilized at around 4.5±0.5 and 3.9±0.2, respectively, for the remainder of the period. Notably, the non-sterilized soil displayed a gradual rise in pH, while the sterilized soil experienced a more abrupt increase (Fig. 1). For both soil conditions, a rapid pH decline to below 3 was observed in the C, F, N, and S-columns contaminated by acidic pollutants on days 0 and 3. Specifically, on day 3, the non-sterilized soil in the C-column showed a minimum pH of 0.76, while the sterilized soil in the N-column exhibited a pH as low as 0.58. Then, both soil conditions displayed a trend of soil pH recovery by day 10. After 10 days, the non-sterilized soil showed average pH values of 4.37, 5.29, 4.56, and 4.34 in the C, F, N, and S-columns, respectively, until the end of the experiment. For the sterilized soil, the C, F, N, and S-columns recorded average pH values of 3.75, 4.23, 3.83, and 3.79, respectively. Both soil conditions exhibited distinct trends in pH recovery after exposure to acidic contaminants. Upon leakage of acidic pollutants, both soil types showed a pH drop to values below 3. These findings suggest that regardless of the presence or absence of soil bacteria, exposure to these contaminants can produce detrimental effects on the soil. The introduction of acidic pollutants leads to a reaction with the soil’s moisture, producing anions and hydrogen ions. This surge in hydrogen ions results in a pH drop and serves as a primary toxicant to bacteria, culminating in a decrease in microbial diversity and negatively impacting the soil [5, 9, 10].
However, during the simulated rainfall period, the trend of pH increase in the soil differed between the two conditions. This suggests that the pH of soil contaminated by acidic pollutants might vary according to the soil’s buffering capacity. In the context of soil, the presence of soil bacteria contributes to nutrient cycling and plays a role in buffering [42]. From days 3 to 10, as the soil’s pH started to recover, the large amount of hydrogen ions supplied by the acidic pollutants bound with the organic matter in the soil, forming organic compounds. These compounds, when utilized by acidophilic bacteria, facilitate the cycling of hydrogen ions, leading to a gradual increase in pH [43, 44]. On the other hand, in sterilized soil where the activity of soil bacteria was eliminated, the pH sharply increased, presumably due to the easier adsorption by rainfall or the leaching away of hydrogen ions [45]. This demonstrates that the presence of bacteria contributing to the soil’s buffering characteristics can be used as an indicator to assess the condition of the soil.
3.2. Bacterial Community Structure3.2.1. Next generation sequencingTo compare the bacterial community structure in the soil at the beginning and end of the acid leak, soil samples were collected from the top layer of the five non-sterilized soils where tap water, HCl, HF, HNO3, and H2SO4 had leaked. Ten samples were collected for 16S rRNA gene amplicon sequencing.
From this analysis, 327,200 high-quality bacterial sequences were obtained, which were further categorized into 6,448 ASVs. Fifteen bacterial species, showing a relative abundance of over 5% at least once in the samples, were identified as the main species (Fig. 2). The top three species in each sample were analyzed and are presented in Table 1. In the initial stages of acidic leakage, species such as Ralstonia syzygii, Citrobacter tructae, Citrobacter werkmanii, Citrobacter cronae, Bacillus clarus, and Rhodococcus qingshengii exhibited high dominance. The average relative abundance of these species in the initial acidic leakage samples was 8.6 ± 10.6%, 6.2 ± 7.0%, 2.8 ± 3.2%, 2.4 ± 2.8%, 8.2 ± 10.5%, and 3.5 ± 7.1%, respectively. After the acidic leakage, Hydrotalea flava, Methylobacterium phyllostachyos, Simkania negevensis, Paraburkholderia terrae, and Tumebacillus ginsengisoli were predominant. In the 34-day post-leakage samples, their average relative abundances were 27.2 ± 8.4%, 11.3 ± 5.9%, 6.4 ± 5.5%, 4.3 ± 8.6%, and 3.1 ± 6.2%, respectively. In the samples of CT1 and NT1 where HCl and HNO3 leaked, an increase in the relative abundance of Ralstonia syzygii, Citrobacter tructae, Citrobacter werkmanii, and Citrobacter cronae was observed. Specifically, in CT1, they showed relative abundances of 10.8%, 10.8%, 4.4%, and 3.9% respectively. In NT1, these percentages were 22.7%, 13.8%, 6.5%, and 5.9% respectively. Notably, in CT1, following the HCl leakage, Rhodococcus qingshengii also held a significant proportion, with 14.2% relative abundance. In FT1 and ST1, where HF and H2SO4 leaked, Bacillus clarus exhibited substantial presence, with relative abundances of 9.4% and 22.7%, respectively.
Following the simulated rainfall, after 34 days, all soil columns showed a high relative abundance of Hydrotalea flava, Methylobacterium phyllostachyos, and Simkania negevensis. Specifically, in CT34, FT34, NT34, and ST34, the relative abundances for Hydrotalea flava were 23.7%, 17.0%, 33.4%, and 34.8%, respectively. Methylobacterium phyllostachyos held considerable proportions, with relative abundances of 10.0%, 3.9%, 13.2%, and 18.1% in these columns. In CT34, where HCl had leaked, Paraburkholderia terrae exhibited a relative abundance of 17.2%. In FT34, affected by the HF leakage, Tumebacillus ginsengisoli held a relative abundance of 12.5%.
In the initial phase of acid leakage, Ralstonia syzygii, known to reduce nitrate to nitrite, displayed a high relative abundance [46], suggesting its prevalence in nitrate-affected soils. Species from the Citrobacter genus also exhibited an elevated relative abundance during this phase. Citrobacter spp. is documented to thrive even in low pH conditions [47]. Specifically, Citrobacter tructae and Citrobacter werkmanii are reported to produce indole related to plant hormones [47, 48]. Citrobacter werkmanii has been recognized for forming robust and stable biofilms. Genetic investigations have identified the presence of genes associated with quorum sensing, such as bsmA, bssR, and bssS, along with a multitude of biofilm-associated genes, including hmsP, tabA, and the csg gene cluster [49, 50]. The formation of such biofilms likely offers protection against physical, chemical, and biological stresses, facilitating attachment to soil surfaces and preventing bacterial washout in dynamic environments, thereby maintaining a high population density. Citrobacter cronae, reported as a close relative to Citrobacter werkmanii, was first identified near the human rectum, indicating its potential to thrive in slightly acidic conditions (pH 5~7) [51]. Rhodococcus qingshengii has been associated with influencing nitrogen cycling in soil environments and is known to assist in the remediation of soils affected by carbendazim [52, 53], which has a reported acidity of pH 4.48. This suggests the bacterium’s ability to persist in low pH environments, potentially accounting for its high relative abundance.
After 34 days, when the soil’s pH had shown considerable recovery, Hydrotalea flava exhibited an elevated relative abundance. This bacterium has been identified in acidic mine drainage, which possesses notably low pH levels, suggesting its adaptability to such conditions [54]. Methylobacterium phyllostachyos, known as a methylotrophic bacterium [55], indicates the production of methyl-related compounds in the soil, signaling ongoing soil remediation. Paraburkholderia terrae has been reported to symbiotically coexist in the roots of leguminous plants alongside Rhizobium and also demonstrates a symbiotic relationship with certain fungi [56, 57]. Its capabilities, such as metabolizing aromatic compounds and fixing nitrogen [58, 59], are indicators of soil health and restoration. Tumebacillus ginsengisoli, characterized by its spore formation and growth at pH 5 [60], is presumed to have accounted for its high relative abundance in soil impacted by HF leakage.
3.2.2. Terminal restriction length polymorphismTo examine the temporal changes in the soil bacterial communities affected by acidic contaminants in the soil, T-RFLP analysis was performed on the 100 collected samples using the 27F and 518R primers and the BsuRI restriction enzyme.
From the analysis, 338 T-RFs were identified using the 27F primer, and 291 T-RFs were identified using the 518R primer, resulting in a total of 629 T-RFs being identified. Notably, using the 27F primer led to the detection of 44 key T-RFs (Fig. 3a) while 27 T-RFs were detected with the 518R primer (Fig. 3b). During the early stages of acid leaking, F205 and R112 showed a high relative abundance. Upon pH recovery of the soil, F212, F252, R113, and R194 exhibited a high relative abundance as well. Intriguingly, some T-RFs were exclusively detected in soils affected by specific acidic contaminants. For instance, T-RFs such as F26, F34, F73, F197, F249, F308, and R319 were observed only in HCl-affected soils. In HF-affected soils, the T-RFs included F54, F202, F293, F304, F305, F306, R11, R53, R54, and R110. For HNO3 affected soils, the T-RFs were F36, F58, F71, F198, F211, F230, R33, R35, and R104, and for H2SO4 affected soils, they were F15, F18, F70, F216, F289, R32, R117, and R198.
During the initial stages of acid leaking, F205 and R112 demonstrated a high relative abundance, and after the acid leakage, F212, F252, R113, and R194 showed a notable relative presence. F205 and R112 could be proposed as early indicators for assessing soil conditions immediately after acid leakage, while F212, F252, R113, and R194 have potential as markers for evaluating soil conditions after pH recovery. Additionally, the unique T-RF patterns, resulting from specific acidic contaminants, suggest the possibility of identifying the respective contaminants.
3.3. Indicator T-RFs for ForensicsTo select statistically significant T-RFs associated with acidic contaminants from the 629 T-RFs identified through T-RFLP experiments, indicator species analysis was conducted using the ‘Indicspecies’ package in R.
Among the 629 T-RFs analyzed, 497 were identified as indicators for acidic pollutants. Specifically, 97 T-RFs were associated with any one of the five acidic pollutants, 44 T-RFs were associated with a combination of two out of the five acidic pollutants, 36 with a combination of three acidic pollutants, and notably, 320 T-RFs were associated with four distinct acidic pollutants (Fig. 4). In soils contaminated with HCl, 11 marker species were identified with F197, R106, F249, and F149 being the most significant. For soil samples exposed to HF, out of the 15 detected species, R52, F263, F293, F305, F234, F199, R54, and F189 were the primary indicators. In the HNO3 contaminated soils, F198 was notably significant among the 8 species. Lastly, in the soils exposed to H2SO4, F259 and F225 were the principal markers out of the 7 identified species. These findings highlight the statistical significant associations of the respective T-RFs.
Despite not having a high relative abundance, specific T-RFs such as R106, F149, R52, F263, F199, F189, F198, F259, and F225 were identified as indicator species. The selection of these T-RFs as indicator species is likely attributed to their heightened sensitivity and representativeness to environmental changes. Some bacteria are recognized as indicator species due to their unique biological traits, like reproductive or metabolic capabilities [61, 62]. Bacteria with high relative abundance might exhibit rapid growth rates because they can metabolize a variety of compounds, including specific substances. In contrast, bacteria with lower relative abundance might grow slower since they only metabolize specific compounds. According to one report, slow-growing bacteria utilize fewer resources, while fast-growing ones thrive due to their ability to use a multitude of resources. Moreover, the bacterial type, such as Gammaproteobacteria, has been reported to influence the rate at which bacteria grow [63].
Based on these findings, it was hypothesized that indicator species with lower relative abundance might reflect more robust representativeness towards specific contaminants than those with higher relative abundance. Particularly, the sensitivity of such indicator species is likely based on the expression of specific genes [64, 65]. Expressions of genes related to enzyme activities are known to occur more rapidly than changes in soil bacterial diversity indicators. Therefore, the development of T-RFLP analysis targeting functional genes responding to acidic stress is required for more accurately identifying indicator species. In soil environments near mines with low pH values, genes related to dehydrogenase, fluorescein activity, and peroxidase serve as bioindicators [66]. In soils contaminated with organic pollutants, the expression of stress proteins like hsp70 and hsp60 is used for biological monitoring against metal and organic soil exposure [64, 67]. By analyzing these sensitive indicator species targeting specific functional genes, more relevant indicator species can be precisely identified.
3.4. Data Augmentation and Machine Learning-based ForensicsTo predict acidic contaminants, four machine learning algorithms – SVM, KNN, RF, and ANN – were used. The input data consisted of 100 samples containing information on 97 T-RFs that are indicator species for a specific contaminant, and an additional 100 samples were doubled using GAN for data augmentation (Fig. 5).
As a result, using the original 100 samples without augmentation, SVM and KNN showed an accuracy of 72%, but ANN demonstrated the highest performance with 88% accuracy (Table 2). On the other hand, when using a total of 200 samples, including both the original 100 samples and the additional 100 augmented samples, all machine learning algorithms improved their accuracy in predicting acidic contaminants. Notably, ANN achieved a high accuracy of up to 98%. When using the non-augmented samples, the recall values, which indicate how well the models correctly identified the actual cases, for H2SO4 in SVM, KNN, RF, and ANN were relatively low at 40, 20, 40, and 80%, respectively. However, a notable improvement was observed when using the augmented samples as input data; the recall for H2SO4 increased to 90, 70, 70, and 90%, respectively. These results suggest that using GAN can amplify the characteristics of specific samples [36, 68].
The advancement of molecular biology equipment has significantly reduced the time and cost of conducting molecular biology experiments [17]. However, obtaining biological da ta still requires intensive time and costs due to complex procedures, including molecular biological monitoring such as T-RFLP, which can take up to a day to process and yield results [69, 70]. Moreover, the rigorous screening process in selecting crucial data, such as indicator species, often reduces the amount of available information. It has also been reported that a small amount of input data is insufficient for building a proper ANN model, leading to research estimating the minimum sample size needed to address this issue [71, 72]. In supervised machine learning algorithms, a small amount of input data often leads to overfitting during the training phase, preventing the model from being perfectly generalized [73, 74]. Therefore, there are many efforts to avoid this through cross-validation, regularization, dimensionality reduction, and data augmentation [75, 76]. Especially, data augmentation can prevent overfitting by increasing the diversity of data, thus allowing the model to be trained to respond to a variety of scenarios [76, 77]. A forensic study processed 50 samples using two restriction enzymes, HhaI and AluI, to generate T-RF data for increased accuracy [78]. Another study in nitrogen treatment bioreactors used the SMOTE (Synthetic Minority Over-Sampling) technique to generate additional data points, resulting in an increase in prediction accuracy from 84% to 88.2% [38]. Therefore, if molecular biological data generated from a limited number of experiments can be meaningfully augmented, it could be a cost-effective method for environmental monitoring.
When comparing machine learning algorithms, both SVM and KNN showed an identical accuracy of 72% in predicting acidic contaminants, which is believed to be due to their methods of learning the relationship between input values and target variables. As the volume of data increases, SVM determines a decision boundary using a hyperplane to separate multiple classes, classifying all data within this boundary into one category (Figs. S1 and S5) [79, 80]. In contrast, the performance of KNN depends on the number of nearest neighbors, K, considered, and increasing K can also increase the computational time (Figs. S2 and S6) [81, 82]. This distinction between the two algorithms suggests that as the quantity of data grows, there will be a difference in performance. In conclusion, setting a decision boundary to classify data within a particular region is anticipated to be more effective for processing large datasets than connecting multiple lines [83]. RF may not always be effective in handling large datasets. Composed of multiple decision trees, RF can produce consistent results regardless of the data volume if there are distinct features in even a small amount of data, allowing for classification based on several questions (Figs. S3 and S7) [84–86]. This suggests that RF can achieve excellent performance with limited data. However, data quality is crucial, and the risk of overfitting should not be overlooked [87, 88]. ANN is based on the principles of biological neural networks, leveraging multiple neural network layers to capture the intricate features and relationships within data (Figs. S4 and S8). Such a structure underscores the superior performance of ANN when juxtaposed with other machine learning algorithms (Table 2). Notably, during the learning process of ANN, back-propagation is utilized to compute the derivative of the loss function, adjusting the weights of each node to minimize errors. Increased computations and iterative error corrections adeptly reflect the complexity and diversity of the data [89, 90]. Hence, when employed with a substantial dataset, ANN has demonstrated an impressive accuracy of up to 98% [72, 91]. When comparing the four machine learning algorithms, it is evident that ANN stands out due to its unique advantages in predicting acidic contaminants. Inspired by the structure of biological neural networks, ANN leverages multiple layers to intricately capture features and relationships within data. This complex structure, combined with its ability to adjust weights through backpropagation and handle the complexity and diversity of large datasets, makes ANN particularly effective for such predictions, as evidenced by its impressive accuracy of up to 98%.
4. ConclusionsThe experiment on acid leakage revealed a rapid decrease in soil pH, and while it recovered over time, sterilized and non-sterilized soils showed different recovery patterns. From the 16S rRNA amplicon sequencing, Ralstonia syzygii and Citrobacter spp. dominated initially, but later, Hydrotalea flava and Methylobacterium phyllostachyos emerged as the primary bacteria. The T-RFLP study unveiled temporal changes in the bacterial community due to acid leakage and suggested the potential of identifying pollutants through unique T-RFs patterns. The dataset was augmented using the indicator species data, bringing the total to 200 samples. Testing with machine learning algorithms, ANN demonstrated the highest accuracy, achieving 88% and 98% with the original and augmented datasets, respectively. These results provide a deeper understanding of the differences among machine learning algorithms and performance variations depending on dataset size.
AcknowledgementsThis work was supported by the 2023 Research Fund (Project Number 1.230055.01) of UNIST (Ulsan National Institute of Science & Technology). National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1C1C1009214). This work was supported by the Technological Innovation R&D Program (S3275603) funded by the Ministry of SMEs and Startups (MSS, Korea).
NotesConflict-of-Interest Statement The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Author Contributions S.P. (Ph.D student): Conceptualization, Investigation, Methodology, Validation, Writing - Original Draft, Visualization. M.T.N. (Ph.D student): Conceptualization, Investigation, Methodology, Validation, Writing - Original Draft, Visualization. J.J. (Ph.D student): Conceptualization, Investigation, Methodology, Validation. H.B. (Professor): Conceptualization, Supervision, Validation, Writing - Original Draft, Writing - Review & Editing, Funding Acquisition. References1. Havugimana E, Bhople BS, Kumar A, Byiringiro E, Mugabo JP, Kumar A. Soil pollution–major sources and types of soil pollutants. ACS ES. T. Eng. 2017;11:53–86.
2. FAO. Global Assessment of Soil Pollution: Report. Rome: FAO; 2021.
3. Montanarella L, Pennock D, Mckenzie N, et al. The Status of the World’s Soil Resources (Technical Summary). Food and agriculture organization of the united nations. 2015;
4. Hyun SP, Shin D, Moon HS, et al. A multidisciplinary assessment of the impact of spilled acids on geoecosystems: an overview. Environ. Sci. Pollut. Res. 2020;27:9803–9817.
https://doi.org/10.1007/s11356-019-07586-6
5. Shin D, Moon HS, Yoon YY, et al. The current status of strong acids production, consumption, and spill cases in Korea. J. Soil Groundw. Environ. 2014;19(6)6–12.
https://doi.org/10.7857/JSGE.2014.19.6.006
6. Yemelin PV, Kudryavtsev SS, Yemelina NK. The methodological approach to environmental risk assessment from man-made emergencies at chemically hazardous sites. Environ. Eng. Res. 2021;26(4)
https://doi.org/10.4491/eer.2020.386
7. Lee J, Park K. Heavy metal distribution in soils from the Maehyang-ri inland shooting range area. J. Korean Soc. Water Environ. 2008;24(4)407–414.
8. Robinson J. Fluorine: its occurrence, analysis, effect on plants, diagnosis and control. 1977.
9. Brewer R, Garber M, Guillemet F, Sutherland F. Effects of accumulated fluoride on yields and fruit quality of Washington navel oranges. Proc. Am. Soc. Hortic Sci. 1967;91:
10. Bruyn JD, Hulsman A. Fluorine injury in cut flowers of gerbera. Bedrijf Sontwikkeling. 1972;3:209–211.
11. Wei Z, Hu X, Li X, et al. The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PloS. One. 2017;12(4)e0174411.
https://doi.org/10.1371/journal.pone.0174411
12. Brock TD, Madigan MT, Martinko JM, Parker J. Brock biology of microorganisms. Upper Saddle River (NJ): Prentice-Hall; 2003.
13. Park JE, Lee BT, Kim BY, Son A. Bacterial community analysis of stabilized soils in proximity to an exhausted mine. Environ. Eng. Res. 2018;23(4)420–429.
https://doi.org/10.4491/eer.2018.040
14. Shin D, Lee Y, Park J, Moon HS, Hyun SP. Soil microbial community responses to acid exposure and neutralization treatment. J. Environ. Manage. 2017;204:383–393.
https://doi.org/10.1016/j.jenvman.2017.09.014
15. van den Heuvel RN, van der Biezen E, Jetten MSM, Hefting MM, Kartal B. Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 2010;12(12)3264–3271.
https://doi.org/10.1111/j.1462-2920.2010.02301.x
16. Filippidou S, Wunderlin T, Junier T, et al. A Combination of Extreme Environmental Conditions Favor the Prevalence of Endospore-Forming Firmicutes. Front. Microbiol. 2016;7:
https://doi.org/10.3389/fmicb.2016.01707
17. Pattnaik P, Jana AM. Microbial forensics: applications in bioterrorism. Environ. Foren. 2005;6:197–204.
https://doi.org/10.1080/15275920590952874
18. Lee J, Park K. Microbial Community in the TPH-Contaminated Aquifer for Hot Air Sparging using Terminal-Restriction Fragment Length Polymorphism. J. Korean Soc. Water Environ. 2008;24(1)19–29.
19. Kim S, Cho C, Lee E. Studies on the chemical accidents of Korea by the statistics and case review. Korean. J. Hazard. Mater. 2017;5(1)50–58.
20. Lee K, Kwon H, Cho S, Kim J, Moon I. Improvements of safety management system in Korean chemical industry after a large chemical accident. J. Loss Prev. Process Ind. 2016;42:6–13.
https://doi.org/10.1016/j.jlp.2015.08.006
21. Liang Y, Lan J. Numerical simulation and prediction of groundwater pollution caused by the leakage of sulphuric acid storage tank with GMS model. IOP Conf. Ser.: Earth Environ. Sci. 2019;
https://doi.org/10.1088/1755-1315/332/2/022055
22. Evans CD, Monteith DT, Fowler D, Cape JN, Brayshaw S. Hydrochloric acid: an overlooked driver of environmental change. Environ. Sci. Technol. 2011;45(5)1887–1894.
https://doi.org/10.1021/es103574u
23. Öberg G, Sandén P. Retention of chloride in soil and cycling of organic matter-bound chlorine. Hydrol. Processes. 2005;19(11)2123–2136.
https://doi.org/10.1002/hyp.5680
24. Bergkvist B, Folkeson L. Soil acidification and element fluxes of a Fagus sylvatica forest as influenced by simulated nitrogen deposition. Wat. Air Soil Pollut. 1992;65:111–133.
25. Currie HA, Perry CC. Silica in plants: biological, biochemical and chemical studies. Ann. Bot. 2007;100(7)1383–1389.
https://doi.org/10.1093/aob/mcm247
26. Jeon I, Nam K. Change in the site density and surface acidity of clay minerals by acid or alkali spills and its effect on pH buffering capacity. Sci. Rep. 2019;9(1)9878.
https://doi.org/10.1038/s41598-019-46175-y
27. Kim SW, Nam SH, Kwak JI, et al. In situ determination of crop productivity in metal-contaminated, remediated, and reclaimed soils: Significance of ecotoxicological data on assessing soil quality. Environ. Eng. Res. 2023;28(6)doi:https://doi.org/10.4491/eer.2022.785
28. Lee JH, Nam KC, Park KS. Soil washing of abandoned mine soils contaminated by heavy metals. J. Korean Soc. Water Environ. 2006;22(5)871–878.
29. Angermueller C, Pärnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12(7)878.
https://doi.org/10.15252/msb.20156651
30. Cho J, Kim H, Gebreselassie AL, Shin D. Deep neural network and random forest classifier for source tracking of chemical leaks using fence monitoring data. J. Loss Prev. Process Ind. 2018;56:548–558.
https://doi.org/10.1016/j.jlp.2018.01.011
31. Shin H, Byun Y, Kang S, et al. Development of water quality prediction model for water treatment plant using artificial intelligence algorithms. Environ. Eng. Res. 2024;29(2)
https://doi.org/10.4491/eer.2023.198
32. Go WS, Yoon CG, Rhee HP, Hwang SJ, Lee SW. A Study on the prediction of BMI (Benthic Macroinvertebrate Index) using Machine Learning Based CFS (Correlation-based Feature Selection) and Random Forest Model. J. Korean Soc. Water Environ. 2019;35(5)425–431.
https://doi.org/10.15681/KSWE.2019.35.5.425
33. Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958;65(6)386.
https://doi.org/10.1037/h0042519
34. Michalski RS, Carbonell JG, Mitchell TM. Machine learning: An artificial intelligence approach. Springer Science & Business Media; 2013.
35. Wang Y. A mathematical introduction to generative adversarial nets (GAN). arXiv preprint arXiv:200900169. 2020.
36. Zhang D, Ma M, Xia L. A comprehensive review on GANs for time-series signals. Neural Comput. Appl. 2022;34(5)3551–3571.
https://doi.org/10.1007/s00521-022-06888-0
37. Li LG, Yin X, Zhang T. Tracking antibiotic resistance gene pollution from different sources using machine-learning classification. Microbiome. 2018;6:1–12.
https://doi.org/10.1186/s40168-018-0480-x
38. Jeon J, Cho K, Kang J, et al. Combined machine learning and biomolecular analysis for stability assessment of anaerobic ammonium oxidation under salt stress. Bioresour. Technol. 2022;355:127206.
https://doi.org/10.1016/j.biortech.2022.127206
39. Jurkevitch E, Pasternak Z. A walk on the dirt: Soil microbial forensics from ecological theory to the crime lab. FEMS Microbiol. Rev. 2021;45(2)fuaa053.
https://doi.org/10.1093/femsre/fuaa053
40. Reysenbach AL, Giver LJ, Wickham GS, Pace NR. ifferential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 1992;58(10)3417–3418.
https://doi.org/10.1128/aem.58.10.3417-3418.1992
41. Vannini C, Rosati G, Verni F, Petroni G. Identification of the bacterial endosymbionts of the marine ciliate Euplotes magnicirratus (Ciliophora, Hypotrichia) and proposal of ‘Candidatus Devosia euplotis’. Int. J. Syst. Evol. Micr. 2004;54:1151–1156.
https://doi.org/10.1099/ijs.0.02759-0
42. Dai Z, Xiong X, Zhu H, et al. Association of biochar properties with changes in soil bacterial, fungal and fauna communities and nutrient cycling processes. Biochar. 2021;3:239–254.
https://doi.org/10.1007/s42773-021-00099-x
43. Qiu X, Zhang Y, Hong H. Classification of acetic acid bacteria and their acid resistant mechanism. Amb Express. 2021;11:1–15.
https://doi.org/10.1186/s13568-021-01189-6
44. Wang C, Cui Y, Qu X. Mechanisms and improvement of acid resistance in lactic acid bacteria. Arch. Microbiol. 2018;200:195–201.
https://doi.org/10.1007/s00203-017-1446-2
45. Farrah H, Pickering WF. Ph Effects in the Adsorption of Heavy-Metal Ions by Clays. Chem. Geol. 1979;25(4)317–326.
https://doi.org/10.1016/0009-2541(79)90063-9
46. Safni I, Cleenwerck I, De Vos P, Fegan M, Sly L, Kappler U. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Micr. 2014;64(Pt_9)3087–3103.
https://doi.org/10.1099/ijs.0.066712-0
47. Jung WJ, Kim HJ, Giri SS, et al. Citrobacter tructae sp. nov. isolated from kidney of diseased rainbow trout (Oncorhynchus mykiss). Microorganisms. 2021;9(2)275.
https://doi.org/10.3390/microorganisms9020275
48. Brenner DJ, Grimont PA, Steigerwalt AG, Fanning G, Ageron E, Riddle CF. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomospecies. Int. J. Syst. Evol. Micr. 1993;43(4)645–658.
https://doi.org/10.1099/00207713-43-4-645
49. Zhou G, Peng H, Wang YS, Huang XM, Xie XB, Shi QS. Complete genome sequence of Citrobacter werkmanii strain BF-6 isolated from industrial putrefaction. Bmc Genomics. 2017;18(1)1–11.
https://doi.org/10.1186/s12864-017-4157-9
50. Zhou G, Li LJ, Shi QS, Ouyang YS, Chen YB, Hu WF. Effects of nutritional and environmental conditions on planktonic growth and biofilm formation of Citrobacter werkmanii BF-6. J. Microbiol. Biotechnol. 2013;23(12)1673–1682.
http://dx.doi.org/10.4014/jmb.1307.07041
51. Oberhettinger P, Schüle L, Marschal M. Description of Citrobacter cronae sp. nov., isolated from human rectal swabs and stool samples. Int. J. Syst. Evol. Micr. 2020;70(5)2998.
https://doi.org/10.1099/ijsem.0.004100
52. Joshi D, Chandra R, Suyal DC, Kumar S, Reeta G. Impacts of bioinoculants Pseudomonas jesenii MP1 and Rhodococcus qingshengii S10107 on chickpea (Cicer arietinum L.) yield and soil nitrogen status. Pedosphere. 2019;29(3)388–399.
https://doi.org/10.1016/S1002-0160(19)60807-6
53. Chuang S, Yang H, Wang X, Xue C, Jiang J, Hong Q. Potential effects of Rhodococcus qingshengii strain djl-6 on the bioremediation of carbendazim-contaminated soil and the assembly of its microbiome. J. Hazard. Mater. 2021;414:125496.
https://doi.org/10.1016/j.jhazmat.2021.125496
54. Medeiros JD, Leite LR, Pylro VS, et al. Single-cell sequencing unveils the lifestyle and CRISPR-based population history of Hydrotalea sp. in acid mine drainage. Mol. Ecol. 2017;26(20)5541–5551.
https://doi.org/10.1111/mec.14294
55. Madhaiyan M, Poonguzhali S. Methylobacterium pseudosasicola sp. nov. and Methylobacterium phyllostachyos sp. nov., isolated from bamboo leaf surfaces. Int. J. Syst. Evol. Micr. 2014;64(Pt_7)2376–2384.
https://doi.org/10.1099/ijs.0.057232-0
56. Pereira-Gómez M, Ríos C, Zabaleta M, et al. Native legumes of the Farrapos protected area in Uruguay establish selective associations with rhizobia in their natural habitat. Soil Biol. Biochem. 2020;148:107854.
https://doi.org/10.1016/j.soilbio.2020.107854
57. Haq IU, Zwahlen RD, Yang P, Van Elsas JD. The response of Paraburkholderia terrae strains to two soil fungi and the potential role of oxalate. Front. Microbiol. 2018;9:989.
https://doi.org/10.3389/fmicb.2018.00989
58. Iwaki H, Hasegawa Y. Degradation of 2-nitrobenzoate by Burkholderia terrae strain KU-15. Biosci. Biotechnol. Biochem. 2007;71(1)145–151.
https://doi.org/10.1271/bbb.60419
59. Yang HC, Im WT, Kim KK, An DS, Lee ST. Burkholderia terrae sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Micr. 2006;56(2)453–457.
https://doi.org/10.1099/ijs.0.63968-0
60. Baek SH, Cui Y, Kim SC. Tumebacillus ginsengisoli sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Micr. 2011;61(7)1715–1719.
https://doi.org/10.1099/ijs.0.023358-0
61. Nguyen J, Lara-Gutiérrez J, Stocker R. Environmental fluctuations and their effects on microbial communities, populations and individuals. Fems. Microbiol. Rev. 2021;45(4)fuaa068.
https://doi.org/10.1093/femsre/fuaa068
62. Scheuerl T, Hopkins M, Nowell RW, Rivett DW, Barraclough TG, Bell T. Bacterial adaptation is constrained in complex communities. Nat. Commun. 2020;11(1)754.
https://doi.org/10.1038/s41467-020-14570-z
63. Kurm V, Van Der Putten WH, De Boer W, Naus-Wiezer S, Hol WG. Low abundant soil bacteria can be metabolically versatile and fast growing. Ecology. 2017;98(2)555–564.
https://doi.org/10.1002/ecy.1670
64. Bhaduri D, Sihi D, Bhowmik A, Verma BC, Munda S, Dari B. A review on effective soil health bio-indicators for ecosystem restoration and sustainability. Front. Microbiol. 2022;13:938481.
https://doi.org/10.3389/fmicb.2022.938481
65. Perez Brandan CG, Huidobro J, Galvan MZ, Vargas Gil S, Meriles JM. Relationship between microbial functions and community structure following agricultural intensification in South American Chaco. Plant Soil Environ. 2016;62:321–328.
https://doi.org/10.17221/19/2016-PSE
66. Masto R, Sheik S, Nehru G, Selvi V, George J, Ram L. Environmental soil quality index and indicators for a coal mining soil. Solid Earth Discuss. 2015;7(1)
https://doi.org/10.5194/sed-7-617-2015
67. Huggett RJ. Biomarkers: biochemical, physiological, and histological markers of anthropogenic stress. CRC Press; 2018.
68. Lin Z, Jain A, Wang C, Fanti G, Sekar V. Using gans for sharing networked time series data: Challenges, initial promise, and open questions. In : Proc. ACM Internet Measurement Conference; 2020; p. 464–483.
https://doi.org/10.1145/3419394.3423643
69. Techtmann SM, Hazen TC. Metagenomic applications in environmental monitoring and bioremediation. J. Ind. Microbiol. Biotechnol. 2016;43(10)1345–1354.
https://doi.org/10.1007/s10295-016-1809-8
70. van Straalen NM, Roelofs D. Genomics technology for assessing soil pollution. J. Biol. 2008;7(6)1–5.
https://doi.org/10.1186/jbiol80
71. Raudys Š, Jain AK. Small sample size problems in designing artificial neural networks. Machine Intelligence and Pattern Recognition. 11:Elsevier; 1991. p. 33–50.
https://doi.org/10.1016/B978-0-444-88740-5.50008-6
72. Prol Castelo G. Minimum sample size estimation in Machine Learning. 2022.
73. Trivedi UB, Bhatt M, Srivastava P. Prevent overfitting problem in machine learning: a case focus on linear regression and logistics regression. Innovations in Information and Communication Technologies (IICT-2020). Advances in Science, Technology & Innovation. Springer; Cham: 2021. p. 345–349.
https://doi.org/10.1007/978-3-030-66218-9_40
74. Salman S, Liu X. Overfitting mechanism and avoidance in deep neural networks. arXiv preprint arXiv:190106566. 2019;
https://doi.org/10.48550/arXiv.1901.06566
75. Ying X. An overview of overfitting and its solutions. In Journal of Physics: Conference Series. 2019;1168(2)22022.
https://doi.org/10.1088/1742-6596/1168/2/022022
76. Santos CFGD, Papa JP. Avoiding overfitting: A survey on regularization methods for convolutional neural networks. ACM Comput. Surv. (CSUR). 2022;54(10s)1–25.
https://doi.org/10.1145/3510413
77. Rebuffi SA, Gowal S, Calian DA, Stimberg F, Wiles O, Mann T. Fixing data augmentation to improve adversarial robustness. arXiv preprint arXiv:210301946. 2021;
https://doi.org/10.48550/arXiv.2103.01946
78. Quaak FC, Kuiper I. Statistical data analysis of bacterial t-RFLP profiles in forensic soil comparisons. Forensic Sci. Int. 2011;210(1–3)96–101.
https://doi.org/10.1016/j.forsciint.2011.02.005
79. Hearst MA, Dumais ST, Osuna E, Platt J, Scholkopf B. Support vector machines. IEEE Intell. Syst. Appl. 1998;13(4)18–28.
https://doi.org/10.1109/5254.708428
80. Dai TT, Dong YS. Introduction of SVM related theory and its application research. In : 2020 3rd International Conference on Advanced Electronic Materials, Computers and Software Engineering, AEMCSE. 2020; IEEE; p. 230–233.
https://doi.org/10.1109/AEMCSE50948.2020.00056
81. Taunk K, De S, Verma S, Swetapadma A. A brief review of nearest neighbor algorithm for learning and classification. In : 2019 International Conference on Intelligent Computing and Control Systems (ICCS); 2019; IEEE; p. 1255–1260.
https://doi.org/10.1109/ICCS45141.2019.9065747
82. Zhang Z. Introduction to machine learning: k-nearest neighbors. Ann. Translational Med. 2016;4(11)
https://doi.org/10.21037/atm.2016.03.37
83. Wiyono S, Abidin T, Wibowo D, Hidayatullah M, Dairoh D. Comparative study of machine learning knn, svm, and decision tree algorithm to predict students performance. Int. J. Res. -Granthaalayah. 2019;7(1)190–196.
https://doi.org/10.29121/granthaalayah.v7.i1.2019.1048
84. Louppe G. Understanding random forests: From theory to practice. arXiv preprint arXiv:14077502. 2014;
https://doi.org/10.48550/arXiv.1407.7502
86. Rokach L, Maimon O. Decision trees. Data Min. Knowl. Discov. Handb., Springer US, Boston, MA. 2005;165–192.
https://doi.org/10.1007/0-387-25465-X_9
87. Han S, Williamson BD, Fong Y. Improving random forest predictions in small datasets from two-phase sampling designs. BMC Med. Inform. Decis. Mak. 2021;21(1)1–9.
https://doi.org/10.1186/s12911-021-01688-3
88. Qi Y. Random forest for bioinformatics. Ensemble Machine Learning: Methods and applications. 2012;307–323.
https://doi.org/10.1007/978-1-4419-9326-7_11
89. Jain AK, Mao J, Mohiuddin KM. Artificial neural networks: A tutorial. Comput. 1996;29(3)31–44.
https://doi.org/10.1109/2.485891
90. Uhrig RE. Introduction to artificial neural networks. In : Proceedings of IECON’95-21st Annual Conference on IEEE Industrial Electronics; 1995; IEEE; p. 33–37.
https://doi.org/10.1109/IECON.1995.483329
91. Raudys SJ, Jain AK. Small sample size effects in statistical pattern recognition: Recommendations for practitioners. IEEE Trans. Pattern Anal. Mach. Intell. 1991;13(3)252–264.
https://doi.org/10.1109/ICPR.1990.118138
Fig. 1Changes in soil pH due to acid leakage and rainfall weathering simulation. a: pH of soil Taqwater leaked. b: pH of soil leaked HCl c: pH of soil from which HF leaked. d: pH of soil from which HNO3 leaked. e: pH of soil from which H2SO4 leaked. 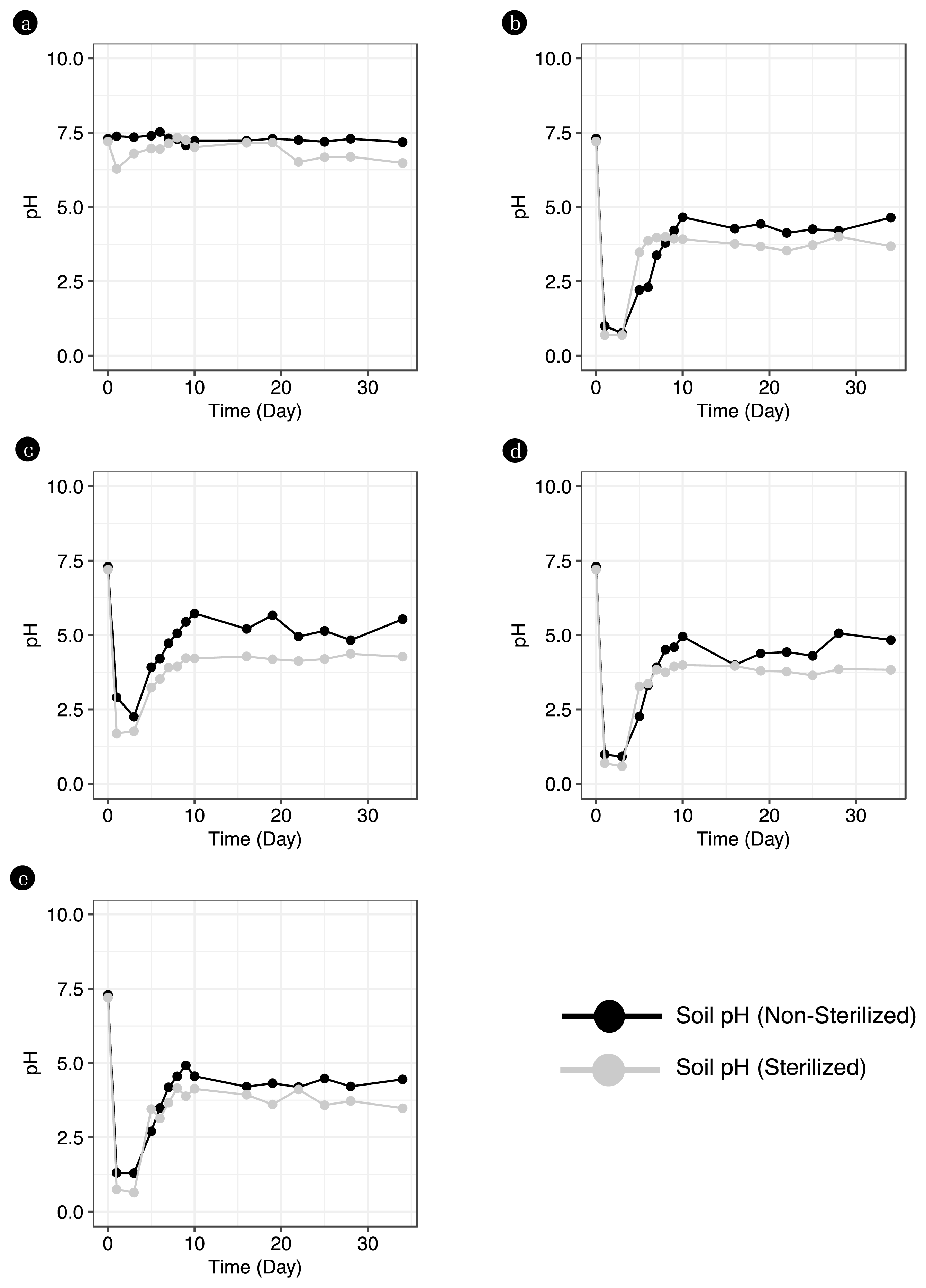
Fig. 2Changes in the bacterial community on the first day and after 34 days of acid leakage using 16S rRNA gene amplicon sequencing. 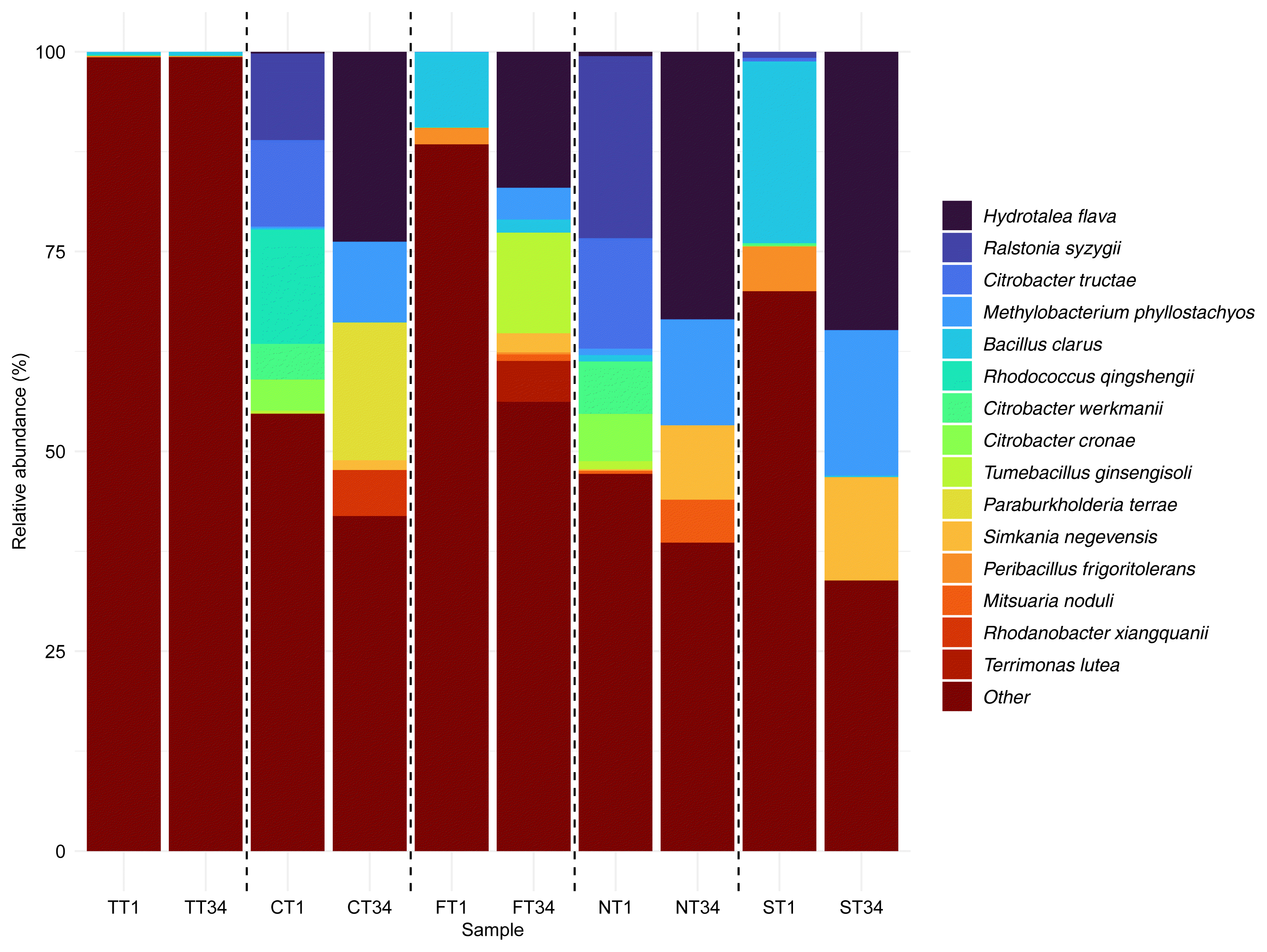
Fig. 3Time series changes in the bacterial community using T-RFLP with a: 27F forward primer and b: 518R reverse primer. 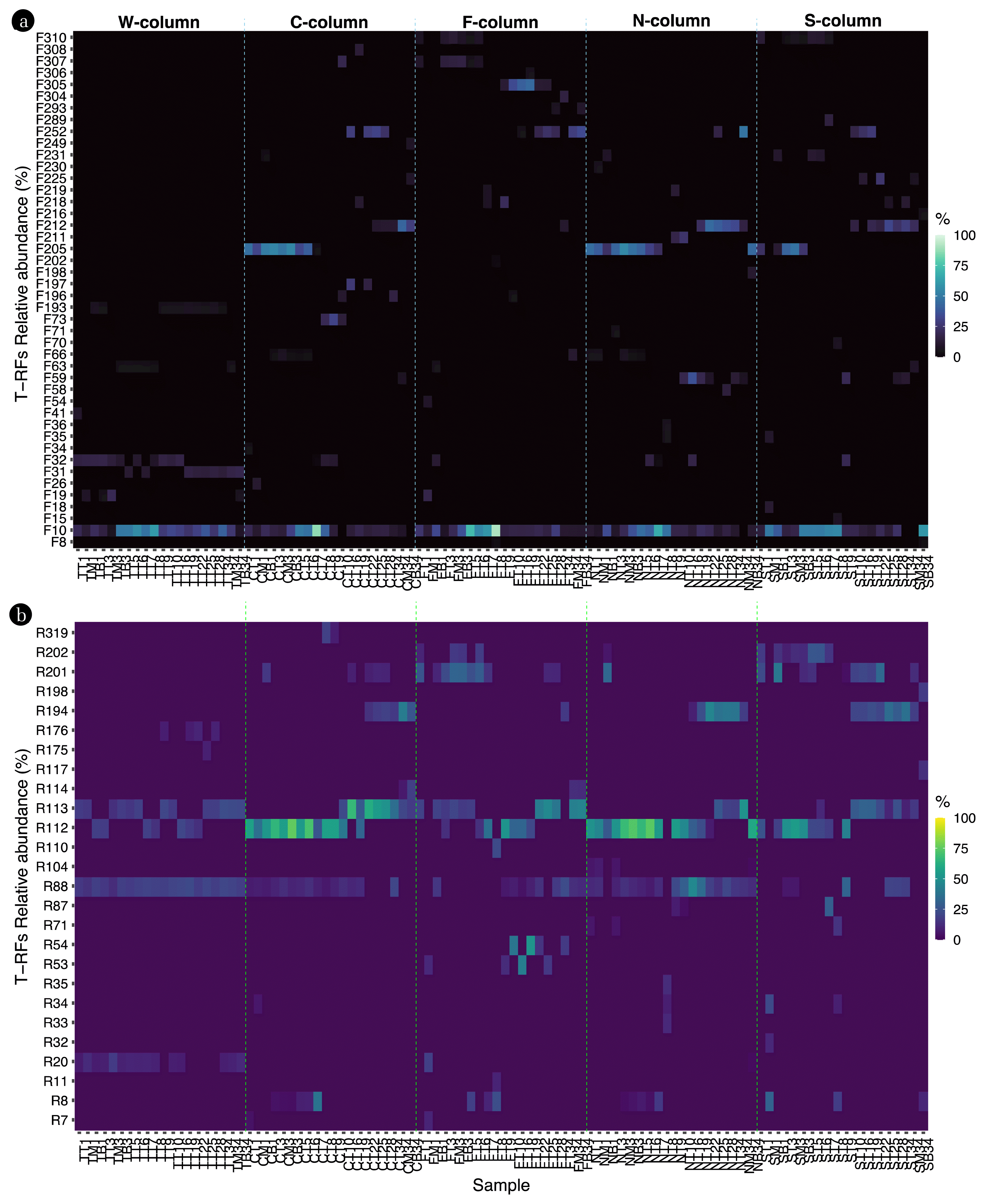
Fig. 5Standardized relative abundance values of 100 samples and 97 selected indicators T-RF. a: Raw sample, b: Augmented sample. 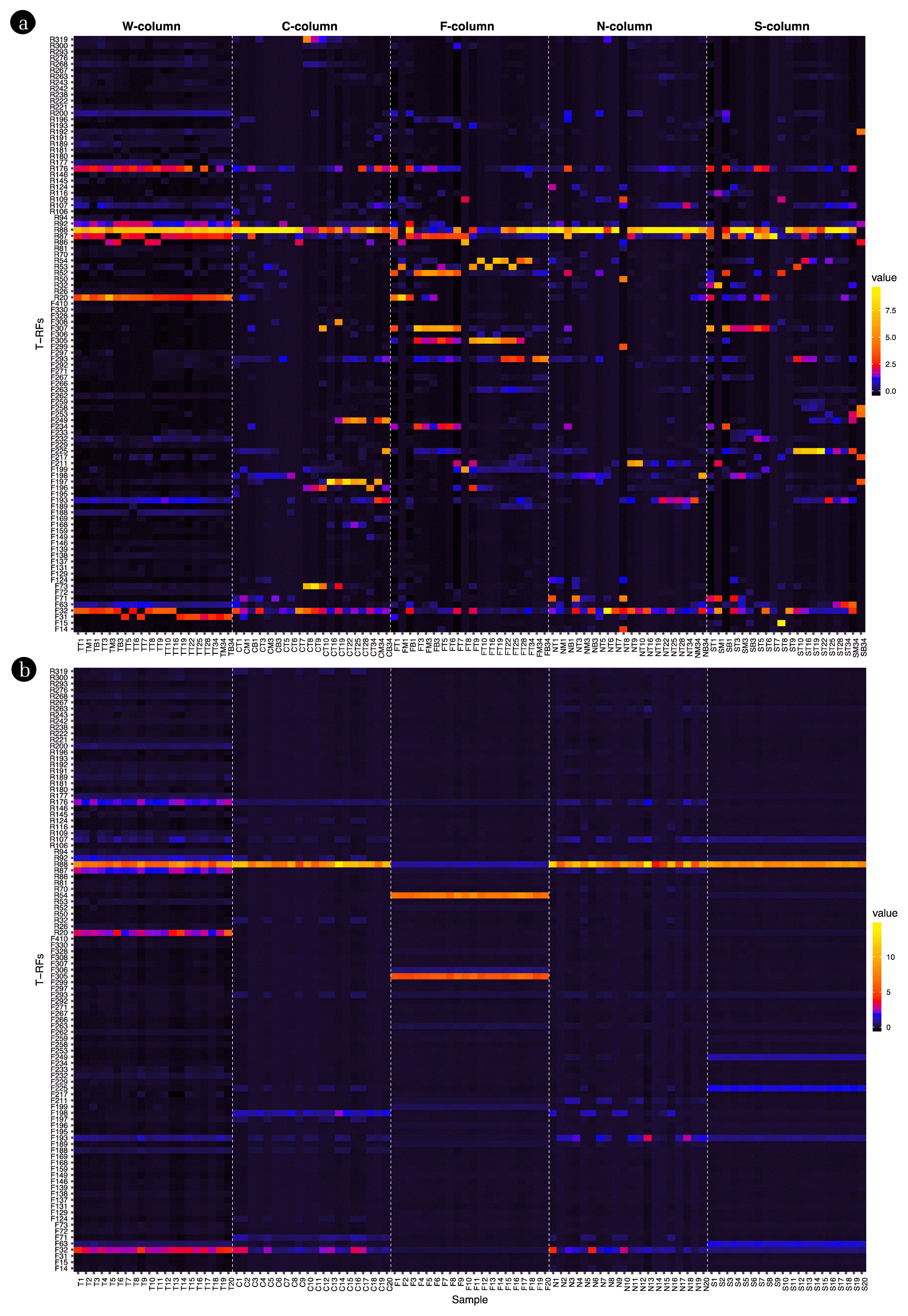
Table 1Taxonomic information of the top 3 bacterial species in soils leaked from contaminants Table 2Classification performance of acidic pollutants by algorithm: Raw sample vs. augmented sample |
|
|||||||||||||||||||||||||||||||||||||||