AbstractNatural organic matter (NOM) is a ubiquitous substance in natural aquatic ecosystems that is a significant component in any experiment involving water. However, experimentally simulating the aquatic conditions using NOMs from natural sources remains difficult. As a result, previous experimental studies have predominantly relied on commercialized NOMs. This study aimed to comprehensively compare the characteristics of two commercialized NOMs (Suwannee River NOM: SRNOM, Mississippi River NOM: MRNOM) and two regional NOMs (Nakdong River NOM: NDNOM, effluent organic matter: EfOM) using various analytical methods during water dissolution. Both commercialized NOMs showed low conductivity (SRNOM: 28.6 μS/cm, MRNOM: 35.4 μS/cm) and were highly humidified (SRNOM: HIX 14.22, MRNOM: HIX 11.44), whereas NOMs from natural water had relatively higher conductivity (NDNOM: 365.7 μS/cm, EfOM: 398.8 μS/cm) and lower humification (NDNOM: HIX 2.47, EfOM: HIX 2.50). The SRNOM and MRNOM contained large amounts of tannin-like substances (SRNOM: 43.5%, MRNOM: 43.1%). The NDNOM had a humidification state similar to that of the EfOM, except for the portion of protein-like biopolymer, which was smaller. The different characteristics of the samples can be critical in selecting appropriate NOMs for use in future studies because they can significantly influence the experimental chemical reaction.
Graphical Abstract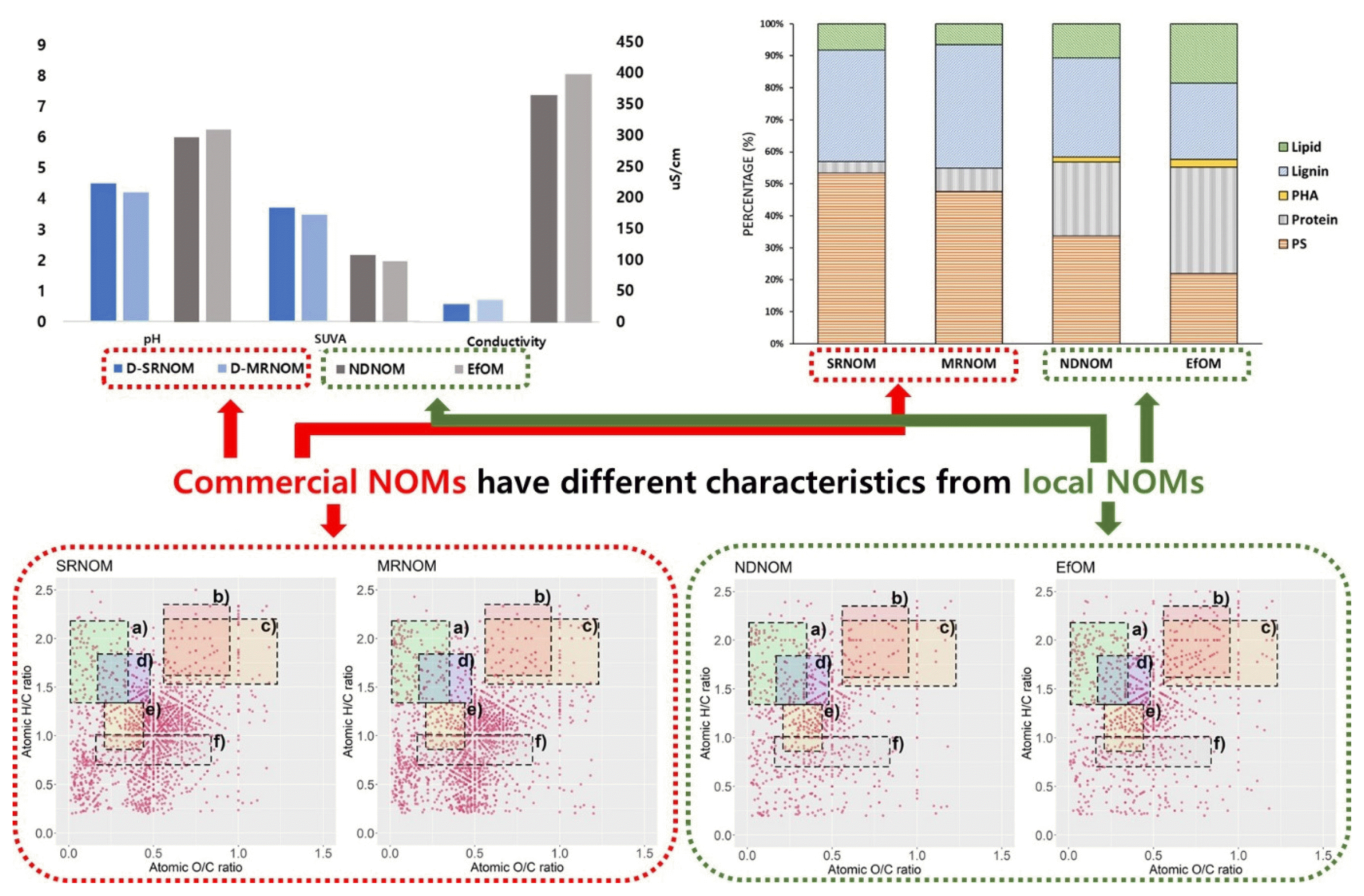
1. IntroductionNatural organic matter (NOM) is a ubiquitous substance in the aquatic environment and is derived from the metabolism and decomposition of living organisms. Since NOM originates from natural sources, its characteristics can vary significantly based on environmental factors, such as climate, source location, land use, and ecosystem dynamics [1–3]. Hence, their chemical characteristics are complex and heterogeneous, and further clarifications are essential to understand their effects in an aquatic environmental system.
NOM consists of carbon-containing substances and energy sources for aquatic organisms and can, thus, influence the carbon cycle and ecosystems. In addition, it is considered a problematic material that causes taste, odor, and membrane fouling and produces disinfection byproducts in drinking water [4–6]. Therefore, NOM has been the subject of extensive research over the past few decades in environmental science and engineering fields.
NOM is classified into humic and non-humic substances. Non-humic substances encompass molecules of biological origin, such as carboxylic, phenolic, and amino acids, lipids, and carbohydrates. Meanwhile, humic substances are further divided into humic acid (HA), fulvic acid (FA), and humins based on their solubility in water at specific pH levels. Specifically, HAs are soluble only in alkaline solutions, FAs are soluble in both acidic and alkaline solutions, and humins are insoluble in both acidic and alkaline solutions.
The most well-known NOM is Suwannee River NOM (SRNOM) in aquatic chemistry studies [7], which is obtained from a blackwater river with a relatively high concentration of dissolved organic carbon concentration (25–75 mgC/L). It comprises allochthonous organic carbons originating from peat and decomposing vegetation [8]. It is hydrophobic and contains an organic matter that has a large molecular weight [9–11]. Since it is commercial NOM, it has been used to investigate the chemical reactions or dynamics of water pollutants.
However, the usage of regional or local natural organic matter (NOM) can show more practical results than commercial NOM when researchers try to design a water treatment process for a specific region. This is because NOM characteristics are affected by their origin.
Malcolm and MacCarthy [12] reported several limitations in using commercial HA in water and soil research. Based on their comparison of the characteristics of five different commercial humic acids using 13C NMR spectroscopy, commercial products were unable to simulate actual soil and water humic substances, as they lacked critical information on the source, extraction, and pre-treatment methods used. In addition, using commercial HA increased the uncertainty of experimental results. Rho et al. [13] also compared the hydrophobicity of SRNOM and NOM obtained from the Han River in South Korea to determine the various organic characteristics. The former contained 97% of hydrophobic fraction, whereas the latter only had 23%. In addition, Watson et al. [5] reported differences in the reactivity of NOMs obtained from various sources. Three drinking water sources in Australia were studied, in which SRNOM was treated using different water purification processes to compare the removal efficiency between the NOMs from the three water sources and SRNOM. The dissolved organic carbon (DOC) removal rate of SRNOM was lower than that of the three Australian samples, owing to the large proportion of NOMs with high molecular weights.
Although reginal or local NOM is preferred in experiments, it is difficult to extract local NOM from natural water owing to their low concentration, which can range between 3–10 mgC/L. Therefore, obtaining a dozen milligrams of NOM powder requires approximately 10 L of water. Moreover, sample loss can become a serious issue during the isolation process owing to the heterogeneity and variety of NOM. Ion exchange is also necessary to remove minerals during NOM concentration. Owing to the complex nature of isolation pre-treatment and the potential loss of NOM during the process, it is challenging to perfectly simulate the actual water conditions using isolated NOM, even when the NOM is effectively isolated. Therefore, selection between commercial and local NOM is essential to design an experiment.
Despite the importance of NOM characterization according to its origin, most studies on NOMs have primarily focused on SRNOM, particularly on the analysis of their optical properties and molecular structure using UV and fluorescence spectroscopy, nuclear magnetic resonance (NMR), and mass spectrometry. [14, 15]. Aldrich HA had also been frequently used in previous studies; however, only a few have determined its comprehensive characteristics [16–18]. Generally, commercial NOMs significantly influence the organic concentration and the ionic components in solvents upon their dissolution. Hence, understanding the changes in water quality following NOM dissolution is essential.
To date, there has been no detailed comparative analysis between the characteristics of Korean NOM and widely used commercial NOM. Thus, we conducted a comprehensive analysis of the characteristics of NOMs in Korean river water and a wastewater treatment plant and compared them with two representative commercial NOM samples (SRNOM, Mississippi River NOM: MRNOM). The analysis was performed using ion chromatography, UV/florescence spectroscopy, py-GCMS, and orbitrap-mass spectrometry, and the results were compared with the commercial NOM analysis results in previous literature to validate the analytical method used in this study. Our findings can serve as a guideline for NOM analysis using state-of-the-art analytical techniques and reveal the differences in the characteristics of NOMs based on their origin and the factors that need to be considered in NOM experiments.
2. Material & Methods2.1. Sample AnalysisSRNOM and MRNOM were purchased from the International Humic Substances Society (IHSS). Samples were obtained in South Korea on October 2021, in which the Nakdong River NOM (NDNOM) was extracted from the Nakdong River near Bonpo Waterfront Park (35.373295N, 128.646821W), and the effluent organic matter (EfOM) was obtained from the Jinhae municipal wastewater treatment plant (35.1436N, 128.69W), South Korea. pH and water conductivity were measured using a multi-meter (Orion 4 star, Thermo, Cleveland, OH, USA). Following sample collection, the samples were filtrated using a 0.45-μm cellulose acetate membrane filter (Whatman Cat No: 7001 0004, D-47mm, Japan) and stored at 4 °C for further analysis. DOC was measured using the total organic carbon (TOC) analyzer (Sievers M9, SUEZ, France). An ultraviolet (UV)-visible spectrometer (UV-1601, Shimadzu, Japan) was used to measure the UV absorbance of the samples at 254 nm (UVA254). The specific UV absorbance (SUVA) value was calculated from the UVA254 and DOC values. The ion concentration was measured using ion chromatography (ICS-90, Dionex, CA, USA). An AS14 column (250 × 4 mm, Dionex, CA, USA) was then used to measure the cation and anion concentrations. A fluorescence spectrophotometer (RF-6000, Shimadzu, Japan) was used to analyze the qualitative oragnic properties of the selected NOMs. The emission spectra range used was between 250–600 nm, in which the excitation spectra ranged from 230–550 nm. The wavelength step of 2 nm was used to draw the excitation-emission matrix (EEM) in the form of three-dimensional (3D) contour plots. The detected intensities of each sample were divided into three regions for characterization of DOM [19–21] protein-like fluorescence (PLF) at λex/λem=270–290/350–365 nm, fulvic-like fluorescence (FLF) at λex/λem=320–340/410–430 nm, and humic-like fluorescence (HLF) at λex/λem=370–390/460–480 nm.
Fluorescence indices, such as humification index (HIX), fluorescence index (FI), and biological index (BIX), were estimated using the following equations, respectively: [22]
2.2. Pyrolysis-Gas Chromatography/Mass SpectrometryThe selected samples were analyzed in triplicates using the pyrolysis gas chromatography-mass spectrometry (Py-GC-MS) system. The samples were concentrated using a rotary evaporator and were then freeze-dried as a pre-treatment. The system was coupled with a curie-point pyrolyzer (JCI-22, JAI, Japan) and GCMS-QP2010 (Shimadzu, Japan). Approximately 0.1 mg of the sample was prepared for pyrolysis in a ferromagnetic foil (Profoil, JAI). The pyrolysis temperature was 590°C, and helium was used as the carrier gas. The initial gas chromatography (GC) oven temperature was heated from 40°C to 300°C. Temperature increase was set at a rate of 7 °C/min. The holding time at the final temperature was 10 min. The DB-5MS (30 m, i.d. 0.25 mm, film thickness 0.50 μm, Agilent, USA) was used to separate volatilized organic compounds (VOCs), using helium as the carrier gas. The ion source was set to a temperature of 210°C. The compounds were ionized at 70 eV and mass analyzed over a range of 30–500 amu. The pyrolyzed fragments were identified using the NIST 14 library as the reference database. Based on the work of Bruchet et al. [23], each sample result was categorized into six groups: polysaccharide (PS), amino sugar (AS), protein, polyhydroxy aromatic compounds (PHA), lignin, and lipid.
2.3. Orbitrap Mass SpectroscopyAs a pre-treatment step for the orbitrap mass spectroscopy analysis, the solid phase extraction (SPE) was conducted to eliminate inorganic salts. The SPE method and orbitrap mass spectroscopy operating conditions followed those of the method described by Baek et al., [24]. NOM was absorbed using the manual packed SPE containing HLB (Oasis, waters, USA), ENV+ (International sorbent technology, UK), Strata X-AW, and X-CW (Phenomenx, UK). Cartridge conditioning was performed using 5 ml and 10 ml of methanol and deionized water, respectively, in which 1 L of the sample was loaded. Thereafter, the absorbed NOM was extracted using 6 ml ethyl acetate/methanol (50:50 v/v) with 0.5 % ammonia and 3 ml of ethyl acetate/methanol (50:50 v/v) with 1.7 % formic acid. This method demonstrated a high recovery rate of the substrate, exceeding 75% [25]. The pretreated sample was analyzed using the Ultimate 300 UPLC system (Thermo Fisher Scientific, San Jose, CA, USA) coupled with the exactive and orbitrap mass spectrometry (Thermo Fisher Scientific, San Jose, CA, USA) with a heated electrospray interface (HESI). HESI was operated under the following conditions: 45 L/min sheath gas flow, 320 capillary temperature, 3800V/3000V spray voltage, 10 arbitrary units auxiliary gas pressure, and 2 arbitrary units ion sweep gas. The injection volume was 200 μl with the methanol mobile phase. Mass spectra were recorded between 100 and 2000 m/z. The molecular formulas were assigned using a compound identification algorithm on MATLAB, as introduced by EB Kujawinski and MD Behn [26].
3. Results and Discussion3.1. NOM Dissolved Water Quality Analysis
Table 1 summarizes the results of the water quality analysis and compares the difference among the dissolved NOM samples. The pH values of the dissolved SRNOM (D-SRNOM) and MRNOM (D-MRNOM) were approximately 4.3, and those of the NDNOM and dissolved EfOM were approximately 7. The low pH of the D-SRNOM and D-MRNOM is attributed to the cation-exchange resin that was used for desalting the NOMs. According to Green et al. [27], IHSS NOM is concentrated using the reverse-osmosis (RO) membrane system. Then, desalting of the concentrated NOM results from the reduction of the high concentration of salts by the ion-exchange resin. With the release of hydrogen ions by the cation exchange resin during cation adsorption from the sample, the functional group of NOM is then protonated [28]. Hence, the low conductivity and low pH of the D-SRNOM and D-MRNOM can be attributed to the desalting process.
The difference in the hydrophobicity of each NOM is represented by the specific UV absorbance (SUVA) value, as high SUVA values correspond to higher hydrophobicity. The D-SRNOM had the highest hydrophobic NOM, followed by D-MRNOM, NDNOM, and EfOM. The ion concentration analysis also showed a similar pattern of conductivity, in which D-SRNOM and D-MRNOM had low concentrations of anions and cations. However, the conductivity of NDNOM and EfOM was higher than that of D-SRNOM and D-MRNOM. This may be attributed to the sample source, as NDNOM originated from natural river water, whereas EfOM was sampled from wastewater treatment effluent. As EfOM also underwent a post-chlorination process, high concentrations of chloride were observed. Hence, the comparison of the water quality of dissolved NOM confirmed that the changing water characteristics should be considered when using a commercial NOM to simulate feedwater conditions.
3.2. Fluorescence Spectrometer Results
Fig. 1 shows the 3D EEM spectra of DOMs, in which the fluorescence spectra provide organic fingerprinting information of a heterogeneous sample. As the 3D EEM spectra can be interpreted using the peaks or regions at different excitation and emission wavelengths related to the chemical composition, it is considered as a non-destructive method. Both D-SRNOM and D-MRNOM showed significant humic-like and fulvic-like fluorescence components based on the obtained HLF (D-SRNOM: 31.0, D-MRNOM: 29.7) and FLF (D-SRNOM: 50.0, D-MRNOM: 45.7) values in Table 2. These indicate that approximately 80% of the detected organic matters are classified as allochthonous, whereas only 20% of the detected organic matters are classified as protein-like substances. NDNOM and EfOM showed higher portions of PLF values than D-SRNOM and D-MRNOM, suggesting that these NOMs are not highly humified and contain a relatively high biodegradable portion in their molecular structures.
Various fluorescence indices can be used to examine differences in humification and precursor organic matter [29, 30]. Higher HIX values indicate a higher degree of humification. Humification refers to the process by which complex organic compounds are transformed into humic substances, which are more stable and resistant to degradation. Higher FI values classify the sample as being of terrestrial origin, as they represent the relative contribution of terrestrial and microbial sources to the DOM. Higher BIX values correspond to recently produced DOM of autochthonous origin. The HIX of D-SRNOM and D-MRNOM was 14.22 and 11.44, respectively, indicating high humidification. This result was consistent with the high SUVA values of D-SRNOM and D-MRNOM in Table 1 (D-SRNOM: 3.71, D-MRNOM: 3.48), including the HLF and FLF values. Additionally, their FI values were also relatively lower than those of EfOM, where the latter showed similar values with NDNOM. EfOM showed relatively higher BIX values than D-SRNOM and D-MRNOM, which indicates that it contains higher fractions of organic matters that originate from microbial activity [31, 32], such as those found in biological wastewater treatment plants, owing to the microbial byproduct-like material that contributes to the high fraction of the organic substances. The high PLF result of EfOM also indicates that EfOM is highly influenced by biological processes.
3.3. Py-GCMS ResultsThe analysis of the biomolecular structure of the selected NOMs using the Py-GCMS showed that SRNOM and MRNOM exhibited similar biomolecular compositions, in which they consisted of polysaccharides (approximately 50%) and lignin (approximately 35%), which accounted for the majority of their structure (Fig. 2). Meanwhile, the portions of protein and lipid accounted for approximately 5% and 7%, respectively (Fig. 2). According to the IHSS, the SRNOM that was extracted from blackwater River containing peat and decomposing vegetation, in which lignin and polysaccharide, which were the most dominant components, derived polyphenols or cellulose. Although the detailed characteristics of the MRNOM is not provided by the IHSS, the result implies that the molecular characteristics of MRNOM and SRMON are similar in this study. This was also consistent with the results of previous studies [33, 34] that reported aromaticity ranging between 20–25% and partition coefficients of 1.29 and 1.49 for SRNOM and MRNOM, respectively.
SUVA was detected at 254 nm when the SRNOM and MRNOM was 4.25 and 4.0, respectively. The FI was also relatively similar to each other, with 1.304 and 1.567 for SRNOM and MRNOM, respectively [35]. These results imply that both NOM are influenced by plant decomposition processes, arising from high lignin and polysaccharide portions and low protein and lipid portions. This can be attributed to the protein and lipid-like biopolymers that derive microbial debris.
Wastewater treatment effluents or enriched microbial environmental samples normally contain high portions of protein [3, 36]. In this study, EfOM showed a relatively high portion of protein percentage (31%). Consistent with previous studies, the result indicates that the major source of EfOM differs from SRNOM and MRNOM. As wastewater treatment plants treat highly biodegradable organic matters such as human feces and sewage, they can be easily decomposed using activated sludge processes. Here, the biopolymer composition of EfOM contained a large portion of microbe, which comes from biopolymers such as proteins and lipids. This finding was also similar to that of a previous study by Park et al. [3] that analyzed the DOM of water from a constructed wetland with a wastewater plant effluent as the influent, in which the NDNOM was composed of 34% polysaccharide, 23% protein, 2% PHA, 30% lignin, and 11% lipid. These portions indicate that the organic characteristics of NDNOM were mixed with previous analytical results from SRNOM, MRNOM, and EfOM, as SRNOM and MRNOM are normally characterized by high percentages of lignin and low percentages of proteins. The EfOM showed the highest portion of protein and lipid among the samples. Hence, the observed proportion of the biopolymers of NDNOM is inferred to be between these normal values, as NDNOM originates from a natural ecosystem that contains various organisms.
3.4. Orbitrap MS ResultsOrbitrap mass spectrometry is one of the most powerful tools that can aid in understanding the complexity of NOMs, as it interprets thousands of molecular compositions in a sample and provides estimated molecular formulas for each composition. The relative number of significant elements, such as carbon, hydrogen, and nitrogen, can then be calculated based on the results. Estimation of the hydrophobicity or classification of the biopolymeric character is also possible, as the typical atomic H/C or O/C ratio of each individual biopolymeric compound can be easily expressed. Several studies have suggested that specific regions of some major biomolecular components can be obtained from the van Krevelen diagram [37, 38]. Since NOM is composed of various organic compounds, the results of mass spectrometry reveal the diversity of molecules present. The number of peaks or spots in the diagram indicates the variety of molecules within a sample. The scattering patterns can vary depending on the origin of the NOM. Therefore, the differences observed among samples provide valuable information for characterizing each individual sample.
Laszakovits and MacKay [39] indicated that each study plotted the van Krevelen diagram, differently. Here, data-based chemical class regions were suggested to be used for van Krevelen diagrams, in which major biomolecules can be divided into six groups according to the H/C and O/C ratio: 1) amino sugar-like substance (1.62 < H/C < 2.35, 0.56 < O/C < 0.95), 2) carbohydrate-like substance (1.53 < H/C < 2.20, 0.56 < O/C < 1.23), 3) lignin-like substance (0.86 < H/C < 1.34, 0.21 < O/C < 0.44), 4) lipid-like substance (1.34 < H/C < 2.18, 0.01 < O/C < 0.35), 5) peptide-like substance (1.33 < H/C < 1.84, 0.17 < O/C < 0.48), 6) tannin-like substance (0.70 < H/C < 1.01, 0.16 < O/C < 0.84). In this study, these values were used to interpret the different NOM samples. The assigned MS sample peaks are described in the van Krevelen diagrams based on the CHON formulae in Fig. 3. Both D-SRNOM and D-MRNOM showed similar graphical patterns, in which the H/C ratio from the 0.8–1.2 range and the O/C ratio from the 0.3–0.6 range are highly concentrated. In these regions, lignin-like and tannin-like substances can be particularly found, which is consistent with the result of a previous study analyzing humic substances [37]. The diagrams of the NDNOM and EfOM also indicate that the lignin-like and peptide-like substances are the major biomolecule classes. However, amino sugar-like substances were also found, particularly in the EfOM samples, where they had the highest content among all the samples, indicating that EfOM contains organic matter from microorganisms, since lipid and amino sugar are included in the major compositions of microbial extracellular polymeric substances [40].
Table 3 summarizes the comparison of the number of biomolecules among the samples. D-SRNOM and D-NRNOM contained the largest portion of tannin-like substances (~40%), indicating that the samples are derived from a terrestrial source, as tannin can exist in leaves [41]. Amino sugar, lipid, and peptide-like substances also had low percentages, suggesting that they are highly humified, owing to the high biodegradable components. EfOM contained the highest percentage of amino sugar-like substances (14.7%) among the samples. The portion of lipid and peptide-like substances related to microbial activities were also relatively higher in EfOM than in the other study samples. Meanwhile, NDNOM showed mixed characteristics between D-SRNOM/D-MRNOM and EfOM, owing to the influence of both microbial and terrestrial sources. The sum of the percentages of the lignin and tannin-like substances is indicative of the influence of a terrestrial source, whereas that of amino sugar, lipid, and peptide-like substances indicates the influence of a bacterial source. The influence of the terrestrial and microbial sources on NDNOM was 46.1% and 40.9%, respectively.
4. ConclusionsIn this study, four different types of NOMs were characterized using comprehensive analyses such as ion chromatography, fluorescence spectrometry, Py-GCMS, and high-resolution orbitrap mass spectrometry to determine the characteristic differences that may influence an experiment. The dissolved form of each sample showed different ionic conditions, in which D-SRNOM and D-MRNOM contained high concentrations of the hydrogen ion (pH 4.5 and pH 4.2) and the lowest concentration of total ions (28.6 μS/cm and 35.4 μS/cm). These findings can be attributed to the cation exchange process performed prior to its isolation. The conductivity values of NDNOM and EfOM were relatively similar; however, EfOM had a higher chloride concentration (80 mg/L) than NDNOM (24.4 mg/L), owing to the chlorination performed for disinfection. The fluorescence spectra result also revealed that D-SRNOM and D-MRNOM are the more highly humified samples (HIX: 14.22 and 11.44) than the others. NDNOM was composed of similarly portioned humic and protein-like substances, whereas EfOM was composed of a high portion of protein-like substances, indicating its source from microbial activities. The Py-GCMS and orbitrap MS results showed different molecular characteristics among the samples. SRNOM and MRNOM had large portions of plant-derived biopolymers, such as polysaccharides and lignin, while EfOM had high percentages of protein-like biopolymers. Moreover, the orbitrap MS also showed similar results with Py-GCMS, in which the two commercial NOMs had high percentages of tannin (SRNOM: 43.5%, MRNOM: 43.1%) and lignin-like substances (SRNOM: 18.8%, MRNOM: 17.4%). The influence of microbial activity was also observed in EfOM.
In general, DOMs that originate from different sources have different characteristics, and the influence of their environment simultaneously affects the quality of the solution in which the OM is dissolved. Hence, understanding NOM characteristics is essential when simulating water samples during experiments.
AcknowledgmentsThis study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1C1C1007350).
References1. Ma H, Allen HE, Yin Y. Characterization of isolated fractions of dissolved organic matter from natural waters and a wastewater effluent. Water Res. 2001;35:985–996.
https://doi.org/10.1016/s0043-1354(00)00350-x
2. Maie N, Scully NM, Pisani O, Jaffé R. Composition of a protein-like fluorophore of dissolved organic matter in coastal wetland and estuarine ecosystems. Water Res. 2007;41:563–570.
https://doi.org/10.1016/j.watres.2006.11.006
3. Park J, Choi M, Cho J, Chon K. Transformation of dissolved organic matter in a constructed wetland: A molecular-level composition analysis using pyrolysis-gas chromatography mass spectrometry. Environ. Eng. Res. 2018;23:390–396.
https://doi.org/10.4491/eer.2018.043
4. Chen B, Nam SN, Westerhoff PK, Krasner SW, Amy G. Fate of effluent organic matter and DBP precursors in an effluent-dominated river: A case study of wastewater impact on downstream water quality. Water Res. 2009;43:1755–1765.
https://doi.org/10.1016/j.watres.2009.01.020
5. Watson K, Farré MJ, Knight N. Comparing three Australian natural organic matter isolates to the Suwannee River standard: reactivity, disinfection by-product yield, and removal by drinking water treatments. Sci. Total Environ. 2019;685:380–391.
Https://doi.org/10.1016/j.scitotenv.2019.05.416
6. Xie P, Ma J, Liu W, Zou J, Yue S, Li X, et al. Removal of 2-MIB and geosmin using UV/persulfate: contributions of hydroxyl and sulfate radicals. Water Res. 2015;69:223–233.
https://doi.org/10.1016/j.watres.2014.11.029
7. Qin L, Huang H. Coupled retention of bovine serum albumin and NOM by porous membranes. Sep. Purif. Technol. 2020;235:116122.
https://doi.org/10.1016/j.seppur.2019.116122
8. Mcknight DM, Thorn KA. Humic Substances in the Suwannee River, Georgia: Interactions, Properties, and Proposed Structures. US Geol. Serv. 1989. 87–557.
9. Jermann D, Pronk W, Boller M, Schäfer AI. The role of NOM fouling for the retention of estradiol and ibuprofen during ultrafiltration. J. Membr. Sci. 2009;329:75–84.
https://doi.org/10.1016/j.memsci.2008.12.016
10. Shim J, Yoon N, Park S, Park J, Son M, Jeong K, et al. Influence of natural organic matter on membrane capacitive deionization performance. Chemosphere. 2021;264:128519.
https://doi.org/10.1016/j.chemosphere.2020.128519
11. Wu FC, Evans RD, Dillon PJ. Separation and characterization of NOM by high-performance liquid chromatography and on-line three-dimensional excitation emission matrix fluorescence detection. Environ. Sci. Technol. 2003;37(16)3687–3693.
https://doi.org/10.1021/es020244e
12. Malcolm RL, MacCarthy P. Limitations in the use of commercial humic acids in water and soil research. Environ. Sci. Technol. 1986;20:904–911.
https://doi.org/10.1021/es00151a009
13. Rho H, Chon K, Park J, Cho J. Rapid and effective isolation of dissolved organic matter using solid-phase extraction cartridges packed with Amberlite XAD 8/4 resins. Water. 2019;11:67.
https://doi.org/10.3390/w11010067
14. Her N, Amy G, McKnight D, Sohn J, Yoon Y. Characterization of DOM as a function of MW by fluorescence EEM and HPLC-SEC using UVA, DOC, and fluorescence detection [doc]. Water Res. 2003;37:4295–4303.
https://doi.org/10.1016/S0043-1354(03)00317-8
15. Huber SA, Balz A, Abert M, Pronk W. Characterisation of aquatic humic and non-humic matter with size-exclusion chromatography–organic carbon detection–organic nitrogen detection (LC-OCD-OND). Water Res. 2011;45:879–885.
https://doi.org/10.1016/j.watres.2010.09.023
16. Janot N, Reiller PE, Zheng X, Croué JP, Benedetti MF. Characterization of humic acid reactivity modifications due to adsorption onto α-Al2O3. Water Res. 2012;46:731–740.
https://doi.org/10.1016/j.watres.2011.11.042
17. Nakashima K, Xing S, Gong Y, Miyajima T. Characterization of humic acids by two-dimensional correlation fluorescence spectroscopy. J. Mol. Struct. 2008;883:155–159.
https://doi.org/10.1016/j.molstruc.2007.11.027
18. Tanaka T. Functional groups and reactivity of size-fractionated Aldrich humic acid. Thermochim. Acta. 2012;532:60–64.
https://doi.org/10.1016/j.tca.2011.12.004
19. Baker A. Fluorescence excitation-emission matrix characterization of some sewage-impacted rivers. Environ. Sci. Technol. 2001;35:948–953.
https://doi.org/10.1021/es000177t
20. Chiu TP, Huang WS, Chen TC, Yeh YL. Fluorescence characteristics of dissolved organic matter (DOM) in percolation water and lateral seepage affected by soil solution (S-S) in a lysimeter test. Sensors. 2019;19:4016.
https://doi.org/10.3390/s19184016
21. Hur J, Park MH, Schlautman MA. Microbial transformation of dissolved leaf litter organic matter and its effects on selected organic matter operational descriptors. Environ. Sci. Technol. 2009;43:2315–2321.
https://doi.org/10.1021/es802773b
22. You Y, Park J, Lee B, Lee S. Analysis of optical properties of organic carbon for real-time monitoring. J. Korean Soc. Water Environ. 2021;37:344–354.
23. Bruchet A, Rousseau C, Mallevialle J. Pyrolysis–GC–MS for investigating high-molecular-weight THM precursors and other refractory organics. J. Am. Water Works Assoc. 1990;82:66–74.
https://doi.org/10.1002/j.1551-8833.1990.tb07022.x
24. Baek SS, Choi Y, Jeon J, Pyo J, Park J, Cho KH. Replacing the internal standard to estimate micropollutants using deep and machine learning. Water Res. 2021;188:116535.
https://doi.org/10.1016/j.watres.2020.116535
25. Kang D, Doudrick K, Park N, Choi Y, Kim K, Jeon J. Identification of transformation products to characterize the ability of a natural wetland to degrade synthetic organic pollutants. Water Res. 2020;187:116425.
https://doi.org/10.1016/j.watres.2020.116425
26. Kujawinski EB, Behn MD. Automated analysis of electrospray ionization Fourier transform ion cyclotron resonance mass spectra of natural organic matter. Anal. Chem. 2006;78:4363–4373.
https://doi.org/10.1021/ac0600306
27. Green NW, McInnis D, Hertkorn N, Maurice PA, Perdue EM. Suwannee River natural organic matter: isolation of the 2R101N reference sample by reverse osmosis. Environ. Eng. Sci. 2015;32(1)38–44.
Https://doi.org/10.1089/ees.2014.0284
28. Serkiz SM, Perdue EM. Isolation of dissolved organic matter from the Suwannee River using reverse osmosis. Water Res. 1990;24:911–916.
https://doi.org/10.1016/0043-1354(90)90142-S
29. Huguet A, Vacher L, Relexans S, Saubusse S, Froidefond JM, Parlanti E. Properties of fluorescent dissolved organic matter in the Gironde Estuary. Org. Geochem. 2009;40:706–719.
https://doi.org/10.1016/j.orggeochem.2009.03.002
30. Ohno T. Fluorescence inner-filtering correction for determining the humification index of dissolved organic matter. Environ. Sci. Technol. 2002;36:742–746.
https://doi.org/10.1021/es0155276
31. Bridgeman J, Baker A, Carliell-Marquet C, Carstea E. Determination of changes in wastewater quality through a treatment works using fluorescence spectroscopy. Environ. Technol. 2013;34:3069–3077.
https://doi.org/10.1080/09593330.2013.803131
32. Carstea EM, Bridgeman J, Baker A, Reynolds DM. Fluorescence spectroscopy for wastewater monitoring: a review. Water Res. 2016;95:205–219.
https://doi.org/10.1016/j.watres.2016.03.021
33. Lv J, Han R, Luo L, Zhang X, Zhang S. A novel strategy to evaluate the aromaticity degree of natural organic matter based on oxidization-induced chemiluminescence. Environ. Sci. Technol. 2020;54:4171–4179.
https://doi.org/10.1021/acs.est.9b07499
34. Fu H, Liu K, Alvarez PJJ, Yin D, Qu X, Zhu D. Quantifying hydrophobicity of natural organic matter using partition coefficients in aqueous two-phase systems. Chemosphere. 2019;218:922–929.
https://doi.org/10.1016/j.chemosphere.2018.11.183
35. Du Z, He Y, Fan J, Fu H, Zheng S, Xu Z, et al. Predicting apparent singlet oxygen quantum yields of dissolved black carbon and humic substances using spectroscopic indices. Chemosphere. 2018;194:405–413.
https://doi.org/10.1016/j.chemosphere.2017.11.172
36. Park J, Cho KH, Ligaray M, Choi M. Organic matter composition of manure and its potential impact on plant growth. Sustainability. 2019;11:2346.
https://doi.org/10.3390/su11082346
37. Kim S, Kramer RW, Hatcher PG. Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the van krevelen diagram. Anal. Chem. 2003;75:5336–5344.
https://doi.org/10.1021/ac034415p
38. Wu Z, Rodgers RP, Marshall AG. Two- and three-dimensional van krevelen diagrams: A graphical analysis complementary to the Kendrick mass plot for sorting elemental compositions of complex organic mixtures based on ultrahigh-resolution broadband Fourier transform ion cyclotron resonance mass measurements. Anal. Chem. 2004;76:2511–2516.
https://doi.org/10.1021/ac0355449
39. Laszakovits JR, MacKay AA. Data-based chemical class regions for van krevelen diagrams. J. Am. Soc. Mass Spectrom. 2021;33:198–202.
https://doi.org/10.1021/jasms.1c00230
40. Perera IA, Abinandan S, Subashchandrabose SR, Venkateswarlu K, Cole N, Naidu R, et al. Extracellular polymeric substances drive symbiotic interactions in bacterial–microalgal consortia. Microb. Ecol. 2022;83:596–607.
https://doi.org/10.1007/s00248-021-01772-1
41. Hagerman AE. Extraction of tannin from fresh and preserved leaves. J. Chem. Ecol. 1988;14(2)453–461.
https://doi.org/10.1007/BF01013897
Fig. 3Van Krevelen diagram of selected NOMs. a) Lipid-like substance, b) Amino sugar-like substance, c) Carbohydrate-like substance, d) Peptide-like substance, e) Lignin-like substance, and f) Tannin-like substance. 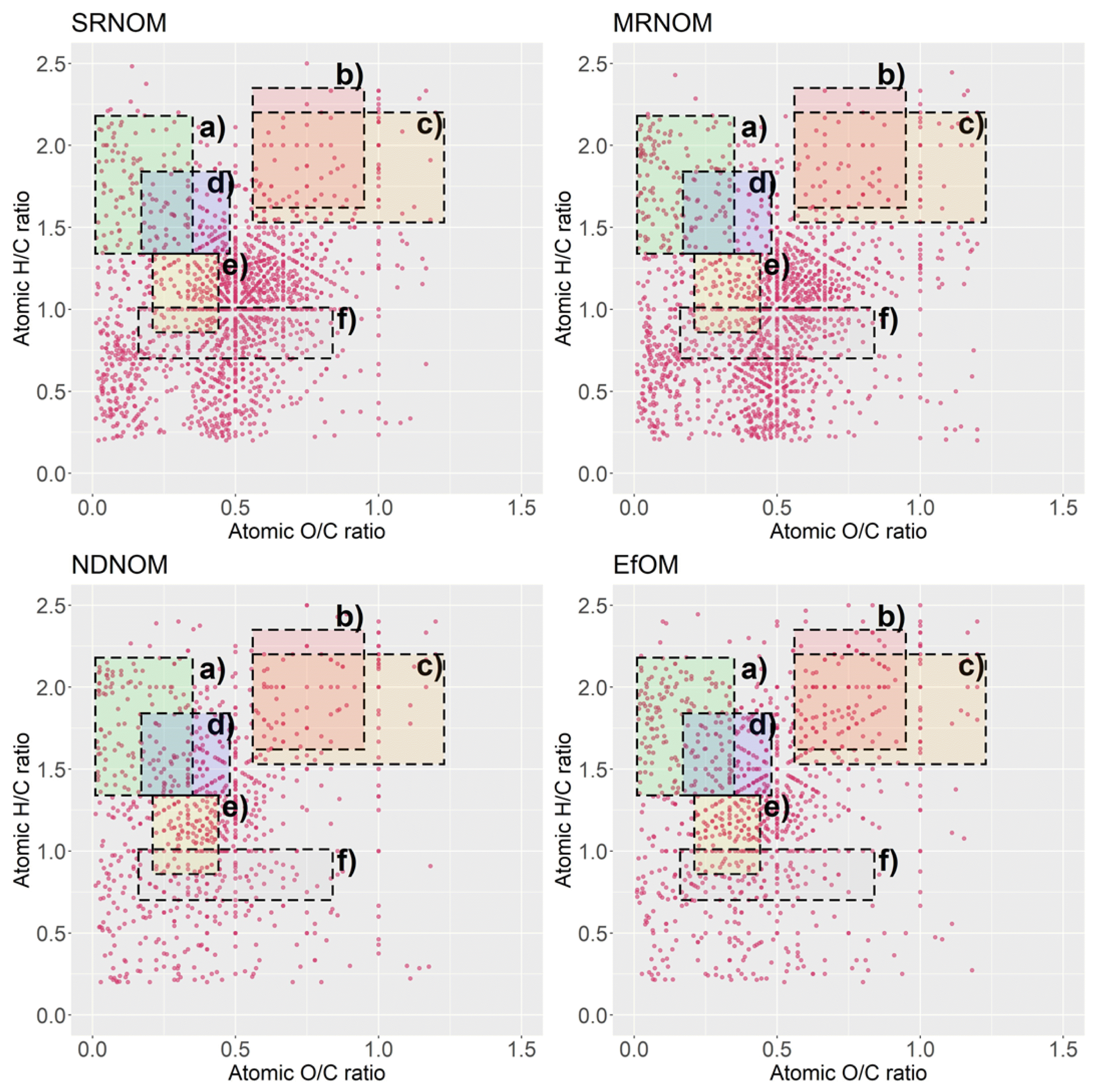
Table 1Water quality analysis results. Table 2Fluorescence indices and organic matter components of NOM samples. Table 3Biopolymer group estimation results from Van Krevelen diagram. |
|
||||||||||||||||||||||||||||||||||