Enhanced Degradation of Sulfamethoxazole Using Immobilized Biomass Reactor
Article information
Abstract
Sulfamethoxazole (SMX) is a common antibiotic frequently detected in the environment. This study investigated enhanced substrate degradation using an immobilized biomass reactor. The degradation rates increased in the batch reactor operation, owing to the increased biomass of immobilized microorganisms in polyurethane foam (PUF). The degradation rate at 50 mg/L was 38.8±3.8 mg SMX/day, and immobilized microorganisms achieved 406.6±42.2 mg dry biomass/g dry PUF under optimum conditions. Two SMX-degrading bacterial strains, Acinetobacter sp. S1 and Pseudomonas sp. S2, were isolated from the immobilized biomass. Furthermore, this study analyzed the possible SMX degradation pathway based on the intermediates of each isolate. The cooperation of these isolated strains resulted in increased degradation and reduced produced metabolites. The degradation by the immobilized cells of the mixed isolated bacteria in continuous operation exceeded 93% SMX at the hydraulic retention time of 36 h at 50 mg/L.
Abstract
Graphical Abstract

1. Introduction
Sulfonamides are antibiotics widely used to prevent and treat bacterial infections in humans and animals. Sulfamethoxazole (4-amino-N-(5-methylisoxazol-3-yl)-benzenesulfonamide, SMX) is a characteristic member of the sulfonamide family used in clinical treatments [1], farming and aquaculture [2]. The antibiotic has been widely detected in environments, that is, streams in the U.S. [3], aquatic environments [4–7], wastewater treatment plants [8], drinking water [9], river water [10], and other water sources [11].
The removal of SMX using conventional wastewater treatment is inefficient [8]. Therefore, biodegradation is considered to be an environmental and effective method to remove the toxic compound. Several microorganisms have been isolated for degrading SMX [12–17], and bacteria can utilize SMX as the sole carbon and nitrogen source [1]. Moreover, the degradation products of these isolated microorganisms have been determined. However, the low removal efficiency and biodegradation of SMX have been reported [12, 18]. In addition, whether pure cultures entirely degrade the antibiotic and whether isolated pure cultures cooperate are still unclear.
SMX degradation using bioreactors has been investigated. For example, bioreactors using freely suspended microorganisms have been applied to degrade antibiotics [19, 20, 21, 22]. In addition, degradations with membrane bioreactors [8, 23] and immobilized cells in bio-carriers in batch reactors [24] have been investigated. The immobilized cell system typically achieves high bioremediation potential because of good biomass retention [25]. Among materials used to immobilize microorganisms, PUF is an effective carrier in removing organic pollutants in a bioreactor [26]. PUF foam has a porous structure, which facilitates microbial growth and colonization and improves the mass transportation of the substrate and nutrients inside the carrier.
This study aims to investigate biomass immobilized in PUF used to degrade SMX in a pilot reactor, isolate SMX-degrading bacteria from the biomass, and determine the degradation pathways of isolates. This study also provides valuable information on improving biodegradation using immobilization biomass in a pilot reactor. Moreover, the degradation pathways and cooperation of isolated bacteria in the degradation of intermediate products were revealed.
2. Materials and Methods
2.1. Chemicals and Media
A mineral salt medium (MSM) with components was prepared, as reported in a previous paper [27]. The MSM consisted of 1.5 g/L Na2HPO4, 0.5 g/L KH2PO4, 0.5 g/L NaCl, 0.2 g/L MgSO4, 0.5 g/L CaCl2, and 1 mL of trace element solution (39.9 mg MnSO4·H2O, 42.8 mg ZnSO4·H2O, 3.8 mg CuSO4·5H2O, 11.6 mg H3BO4, and 27.8 mg FeSO4·7H2O per liter). Ammonium sulfate (0.5 g/L) and succinate (0.5 g/L) were used as the nitrogen and carbon sources, respectively. The pH was adjusted to 7.0±0.1 before sterilizing at 121 °C for 15 min. A solid medium was obtained by adding 2% (w/v) agar for bacterial cultivation. SMX, 3-amino-5-methylisoxazole, and 4-aminothiophenol were separately diluted in ethanol (0.1 M) as stock solutions. All chemicals (microbiology grade) were bought from Sigma-Aldrich (USA) or Merck (Germany).
2.2. Collection of Soil Samples and Enrichment Process
Soil samples near the wastewater effluent from a pharmacy company in Cao Lanh City, Dong Thap Province, Vietnam, were collected and transferred to a laboratory within several hours. The soil samples were smashed, mixed, and sieved through a sieve with an aperture of 2.0 mm diameter. Soil (5 g) was added to a flask containing 100 mL MSM; 1 mg/L SMX was first added to the modified soil and incubated for five days, and another 5 mg/L was added for another five days. Finally, 10 mg/L of the substrate was added every five days for two months. Subsequently, the enrichment medium (1.0 mL) was inoculated into a flask containing 100 mL MSM supplemented with both ammonium sulfate and succinate. Next, SMX was added to the medium at 5 mg/L. The flask was incubated for two days, and the spent medium was used to inoculate to a packed bed bioreactor. The enrichment process was conducted using a shaker at 150 rpm, room temperature (~30 °C), in a dark condition.
2.3. Packed Bed Bioreactor
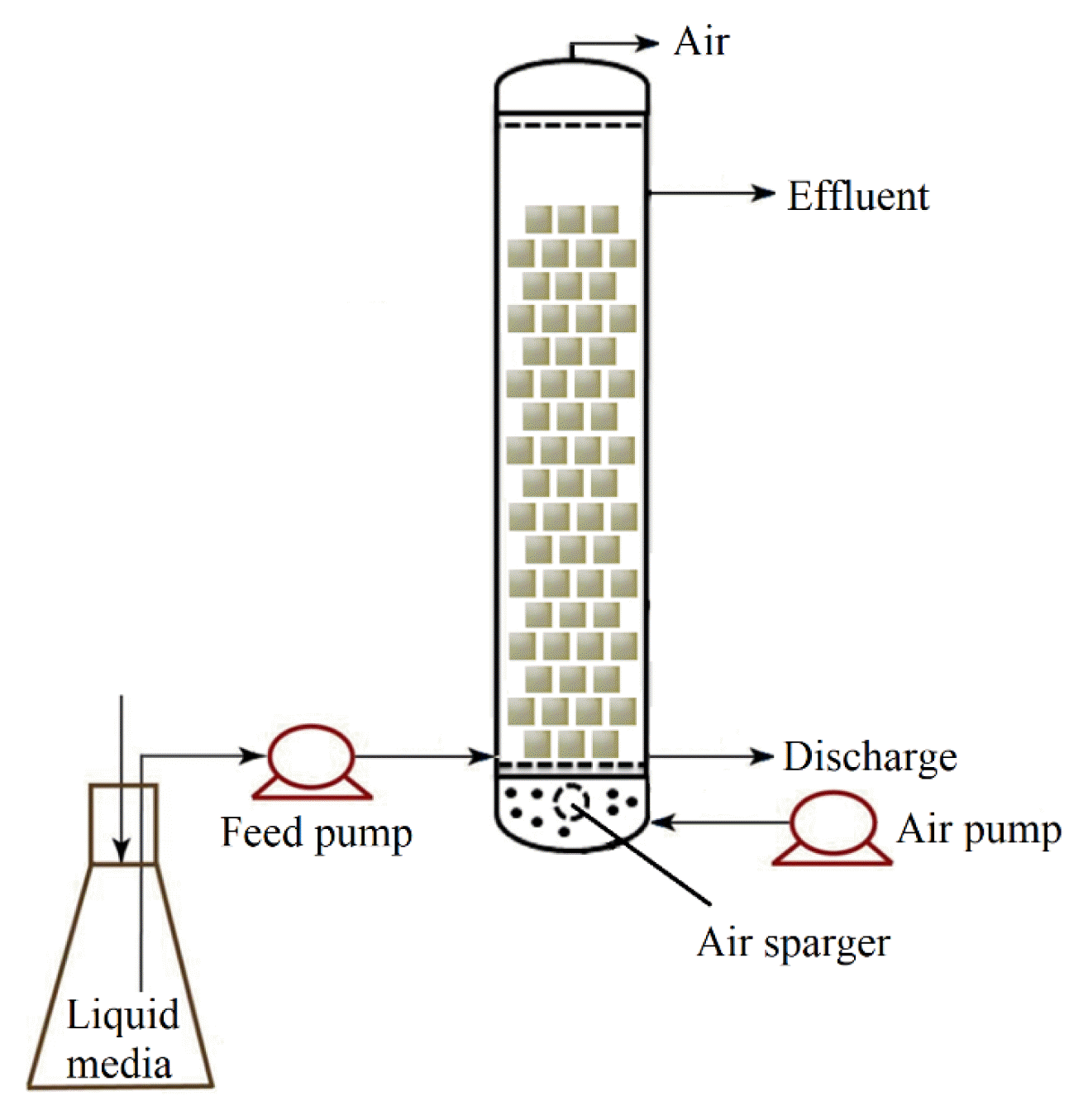
A cylindrical glass column with the following dimensions was fabricated: height = 30 cm, diameter = 5.0 cm, and active bed volume = 500 mL (Fig. 1). PUF with a mean dry weight of 0.0488 g/cm3 and a mean wet weight of 0.277 g/cm3 was used. Three hundred sterilized cubes (1 cm × 1 cm × 1 cm each) of PUF were filled in the reactor. The reactor was connected to a feed tank from which the medium was fed into the column using a peristaltic pump. An air pump was connected to an air sparger at the bottom. The inlet site was set up at the highest liquid level used to discharge under continuous operation. The airflow was controlled at 0.5 L/h in all the experiments.
2.4. Sulfamethoxazole Degradation in Packed Bed Bioreactor
Sterilized MSM medium containing ammonium sulfate and succinate was pumped up to 500 mL into the reactor. As described above, the spent degraded media in the flask were inoculated in the feed tank at 0.5% (v/v) before the first cycle. The degradation was first conducted in a batch operation and then continuously. For the batch operation, the operations at 10, then 25 mg/L SMX were conducted in batch cultures (24 h each) until complete or almost complete removal, and finally at 50 mg/L until the degradation stopped increasing. The reactor was then operated continuously at 50 mg/L SMX with hydraulic retention times (HRTs) of 12, 24, and 36 h.
2.5. Isolation of SMX-degrading Bacteria and SMX Degradation by Bacterial Isolates
A PUF cube containing immobilized bacteria was transferred to a flask containing 100 mL MSM supplemented with 50 mg/L SMX. The flask was shaken at 500 rpm for 6 h at 30 °C. The liquid media were diluted and spread onto solid MSM containing 50 mg/L SMX. Single colonies were purified and subjected to tests to determine their degrading capacities. The method for identifying the obtained bacterial strain was described by Ha and Nguyen [28].
Each isolated bacterial strain was cultured in liquid Luria–Bertani broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) for 24 h. Firstly, the media with bacteria were used for inoculation, from which the bacterial inoculum was transferred to the respective media to obtain an original cell number of approximately 106 CFUs/mL. Next, each strain was inoculated to 0.5 × 106 CFUs/mL MSM for degradation by a mixed culture. Finally, the individual and mixed cultures of isolated strains were determined for degrading in flasks and the reactor.
The degradation in flasks and the reactor was carried out in the MSM medium containing 50 mg/L SMX. For the experiments using flasks, bacteria utilized SMX as the sole carbon and nitrogen source or with ammonium sulfate and succinate in the medium. Meanwhile, the degradation in the reactor was conducted with the addition of ammonium sulfate and succinate. The degradation continued in batch operations until the biomass in the PUF reached the maximum level. The reactor was continuously operated at HRTs of 12, 24, and 36 h. Moreover, the degradation was also conducted from 0.01 to 5.0 mg/L SMX.
2.6. Analytical Methods
The concentrations of the SMX and its metabolites were determined using HPLC, as described by Ha and Nguyen [28]. Liquid media collected from freely suspended cells cultured in the MSM medium with ammonium sulfate and succinate were used to determine the degradation metabolites. The degradation products were analyzed using the gas chromatography–mass spectrometry (GC–MS, Finnigan-MAT MD8000, San Jose, CA, USA) equipped with a DB-5 capillary column (30 m × 0.5 mm × 0.25 m), in which helium was used as a carrier gas with a flow rate of 1 mL/min. The applied oven temperature program was set up at 80 °C for 5 min, then raised 15 °C per min to 260 °C and held there for 3 min. The MS was equipped with an electron ionization source of 70 eV energy. The scanning intervals were 1.5 times/s, and the mass ranged from 30 to 220 m/z. Samples were injected into the GC-MS instrument at a 1:5 split ratio. The determination of enzymes involving ortho- and meta-cleavage pathways was conducted, as described in a previous study [28]. Catechol 1,2-dioxygenase activity was assayed spectrophotometrically by measuring the formation of cis, cis-muconic acid of catechol at 260 nm. In comparison, catechol 2,3-dioxygenase activity was assayed following the formation of 2-hydroxymuconic semialdehyde at 375 nm. The biomass immobilized in PUF was measured based on the different weights before and after the experiments.
2.7. Statistical Analysis
All data obtained from at least three experimental replicates are shown as mean ± standard deviation. Significant differences among means were statistically analyzed using one-way Duncan’s test (p < 0.05) in SPSS program version 22.0.
3. Results and Discussion
3.1. Sulfamethoxazole Degradation in Packed Bed Bioreactor in Batch Operation
The degradation rates and biomass immobilized in the PUF increased with time during the degradation processes (Fig. 2). At 10 mg/L SMX, the degradation was 12.2±3.1% (1.2±0.3 mg/day) in the first day, increased to nearly 100% (9.7±0.1 mg/day) in the seventh day (Fig. 2a). The immobilized microorganisms also increased from 55.3±13.2 mg dry biomass/g dry PUF at the end of the first cycle to 242.1±32.4 mg dry biomass/g dry PUF at the seventh cycle. For the abiotic control at 10 mg/L, PUF absorbed almost 11.4% in the first cycle, 5.5% in the second cycle, and negligible amounts in subsequent cycles.
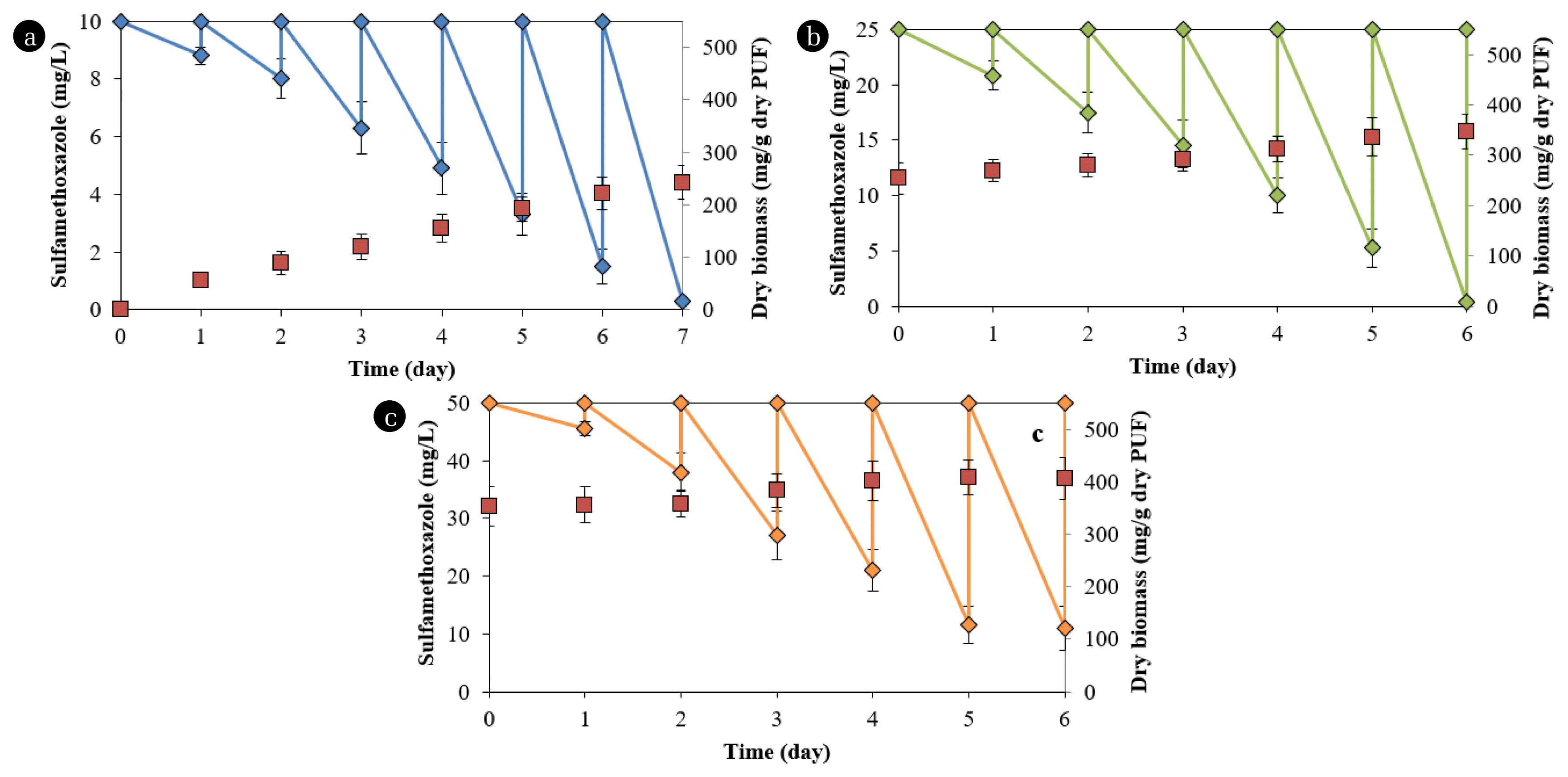
SMX degradation (diamond symbol) by microorganisms immobilized in PUF (square symbol) in reactor at (a) 10, (b) 25, and (c) 50 mg/L SMX. The degradation was conducted in MSM medium supplemented with ammonium sulfate and succinate.
At 25 mg/L, the degradation increased from 28.6±5.2% (7.2±1.3 mg/day) in the first cycle to 98.4±0.8% (24.6±1.0 mg/day) in the sixth cycle (Fig. 2b). The immobilized microorganisms also increased to 347.1±35.5 mg dry biomass/g dry PUF at the sixth cycle for this chemical concentration. The corresponding data for degradation at 50 mg/L were 17.6±4.4% (4.5±0.1 mg/day) and 78.1±7.5% (38.8±3.8 mg/day). The immobilized microorganisms were 406.6±42.2 mg dry biomass/g dry PUF at 50 mg/L SMX (Fig. 2c). The degradation and biomass at the fifth and sixth cycles at 50 mg/L in PUF were not significantly different, indicating that the maximum immobilized biomass was achieved.
The degradation rates in the first cycles of 25 and 50 mg/L were lower than those in the last cycles for 10 and 25 mg/L, respectively. The biomass contents in PUF from the beginning to the second cycle in the reactor with 50 mg/L SMX did not increase. These phenomena probably occurred because the microorganisms required more time to adapt to higher chemical concentrations. Drillia et al. [19] found that SMX was not degraded in the first several days of operation in a sequencing batch reactor, but subsequently, the removal efficiency increased. The removal rates of SMX in a bioreactor are enhanced when the substrate is added after six periods [22]. The amount of immobilized biomass in this study was significantly higher than that in PUF in aerated filter reactors (343 mg dry cell/g dry PUF) [29] but lower than that in the cells in PUF in Erlenmeyer flasks (488 mg dry cell/g dry PUF) used for oilfield degradation [30].
3.2. Isolation and Identification of SMX-degrading Bacteria
Two SMX-degrading bacterial strains were isolated from biomass in PUF. These isolates are named S1 and S2 and have 1,415 and 1,446 pb in 16S rRNA sequences, respectively. A comparison between these sequences with other 16S rRNA sequences available in the NCBI and Ribosomal Database Project showed that S2 and S1 were Acinetobacter sp. and Pseudomonas sp., respectively. Therefore, the isolated strains are Acinetobacter sp. S1 and Pseudomonas sp. S2. The sequences of the isolates have been deposited in GenBank with accession numbers ON505833 and ON505832. In addition, these isolates have been deposited at the Center for Biochemical Analysis (Dong Thap University, Vietnam) with deposition numbers KT2021-S2 and KT2021-S1, respectively.
Acinetobacter and Pseudomonas are the main bacteria that degrade sulfonamides in activated sludge [22]. Some bacterial strains, such as Pseudomonas psychrophila HA-4 [14], Acinetobacter sp. [17], and Sphingobacterium mizutaii LLE5 [31], which utilized SMX as the sole carbon source, have been reported. Achromobacter sp. JL9 utilized SMX as the sole nitrogen source [32]. Some pure cultures, Pseudomonas, Brevundimonas, and Variovorax, were the first to utilize the substrate as the sole carbon and nitrogen source for growth [13]. Other microorganisms have been isolated for degrading SMX, such as some pure cultures [12, 13], Achromobacter denitrificans PR1 [15], and Navicula sp. [16].
3.3. Sulfamethoxazole Degradation by Individual and Mixed Cultures of Isolated Bacteria
Acinetobacter sp. S1 exhibited low utilization of SMX as the sole carbon and nitrogen source (Fig. 3) and did not grow in media without adding any carbon and nitrogen sources. Pseudomonas sp. S2 utilized SMX as the sole carbon and nitrogen source with 31.2±6.8% at 50 mg/L SMX after 72 h (Fig. 3). The substrate degradation was enhanced when ammonium sulfate or both ammonium sulfate and succinate were supplemented. For example, the degradation by Pseudomonas sp. S2 increased by 42.1% with the addition of ammonium sulfate and 53.3% with the addition of ammonium sulfate and succinate in the medium. The degradation of SMX by Acinetobacter sp. S1 was lower than that of Pseudomonas sp. S2 by 35.2% (on average) in the medium supplemented with ammonium sulfate and 27.1% (on average) supplemented with ammonium sulfate and succinate. The degradation percentages of the mixed culture were not statistically different from that degraded by Pseudomonas sp. S2. The presence of ammonium sulfate and succinate stimulated bacterial growth. The growth was higher in media with higher degradation (data not shown). The long-term contact with the antibiotic, both strains could adapt to high concentrations and utilize the substrate to grow. The addition of cosubstrates improved the degradation rates, owing to the increase in biomass, also described in previous studies [32, 33].
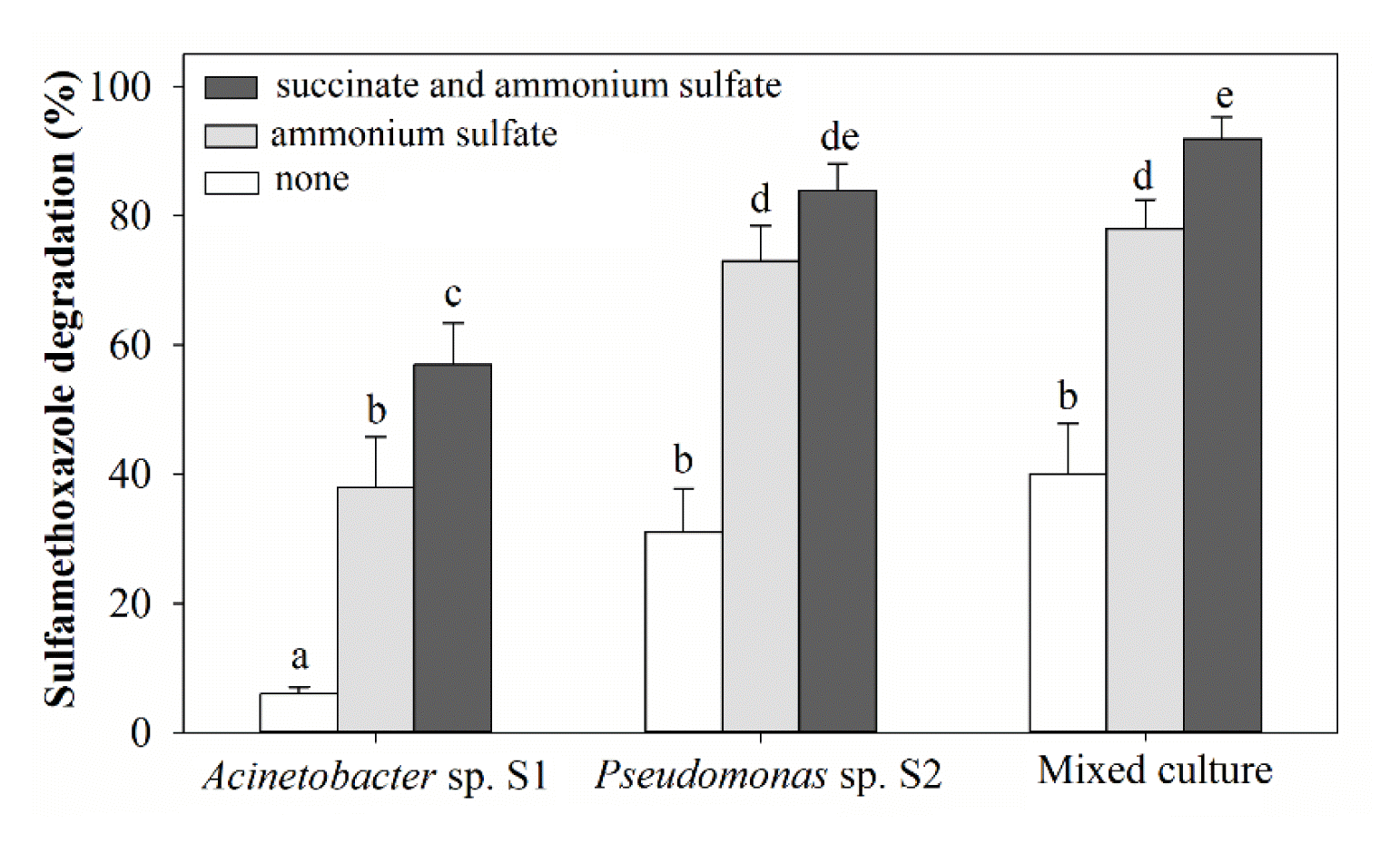
SMX degradation by suspended individual and mixed cultures of Acinetobacter sp. S1 and Pseudomonas sp. S2 cultured for 72 h. The degradation (at 50 mg/L SMX) was conducted in MSM medium with and without supplemental ammonium sulfate and succinate. The error bars indicate the standard deviation for at least three replicates, and different letters in lower case (a–e) represent significant differences among treatments (p < 0.05).
The degradation at small concentrations of SMX, which may be found in environments, was also evaluated. The mixed culture nearly completely utilized the antibiotic at 0.01–5.0 mg/L within 6 to 48 h (Fig. S1) as a sole carbon and nitrogen source. The abiotic control showed no reduction of SMX.
3.4. Sulfamethoxazole Degradation by Mixed Culture of Isolated Bacteria Immobilized in PUF in Packed Bed Bioreactor in Batch Operation
The degradation was conducted in a batch reactor at 50 mg/L SMX. The degradation and bacterial biomass immobilized in the PUF in the rector increased with the number of cycles (Fig. 4). The degradation was 10.9±2.4% (5.5±1.2 mg/day) in the first cycle, increasing to 61.9±7.2% (31.0±3.6 mg/day) in the ninth cycle. The dry biomasses of bacteria immobilized in PUF at the end of the first and eighth cycles were 69.4±7.6 and 355.4±40.2 mg/g dry PUF, respectively. The degradation and bacterial biomass at the eighth and ninth cycles were not statistically different (p<0.05), indicating that the immobilization was saturated.
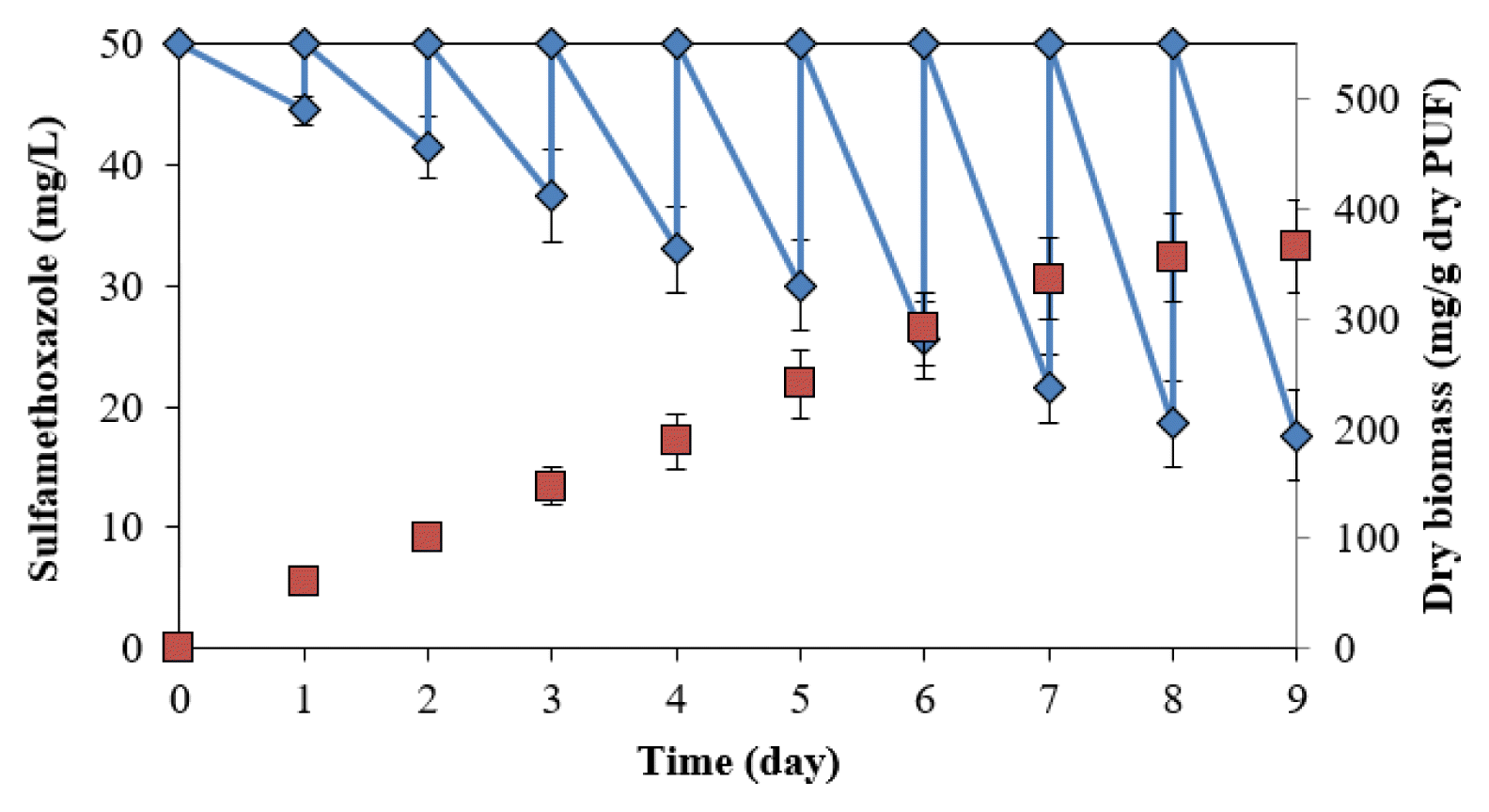
SMX degradation by immobilized cells of Acinetobacter sp. S1 and Pseudomonas sp. S2 in packed bed reactor in batch operation.
The degradation in the packed bed reactor in this study showed higher effective degradation rates than those in reactors in previous studies. For example, batch tests showed limited removal of the antibiotic with < 20% biodegradation after 48 h in a membrane bioreactor at a low concentration (3 μg/L) [23]. Bacteria immobilized in a bio-carrier degraded 59% of the substrate at 100 μg/L after 14 d [24]. SMX (1.0 mg/L) was degraded at 46% in the conventional activated sludge (HRT = 11.5 h) and 52%–55% in membrane reactors (HRTs = 7.2–20 h) [20]. SMX (0.23–0.57 μg/L) was eliminated by approximately 60% in a fixed-bed reactor, compared to approx imately 80% in a membrane bioreactor [8]. The pH was adjusted to neutral, in which SMX is negatively charged, and the pKa2 acts on the sulfanilamide groups protonation (−SO2NH−) [34]. The pH > 5.7 (pKa2), the main form of sulfamethoxazole in water, is ionic, resulting in an increase of solubility in water, which also affects the bioavailability for biodegradation.
3.5. Sulfamethoxazole Degradation in Packed Bed Bioreactor in Continuous Operation
The continuous operation at 12- and 24-h HRTs of a bioreactor demonstrates the actual application. After the PUF immobilization reached maximal levels, degradation in the reactor was performed continuously at 50 mg/L SMX. As described above, the degradation percentages of microorganisms immobilized in PUF were significantly higher than those of the mixed culture of Acinetobacter sp. S1 and Pseudomonas sp. S2 at HRTs of 12 and 24 h. This result indicated that other SMX-degrading microorganisms in the immobilized biomass were not isolated. However, the degradation rates were not statistically significant when the reactor was operated at an HRT of 36 h, with more than 93% SMX removed at 50 mg/L of the substrate (Fig. 5).

Degradation of SMX (50 mg/L) by biomass of nonisolated microorganisms and mixed culture of Acinetobacter sp. S1 and Pseudomonas sp. S2 immobilized in PUF. The degradation was conducted in the reactor in continuous operation at HRTs of 12, 24, and 24 h. The error bars indicate the standard deviation for at least three replicates, and different letters in lower case (a–e) represent significant differences among treatments (p < 0.05).
3.6. Identification of Metabolites and Degradation of Metabolites by Isolated Bacteria
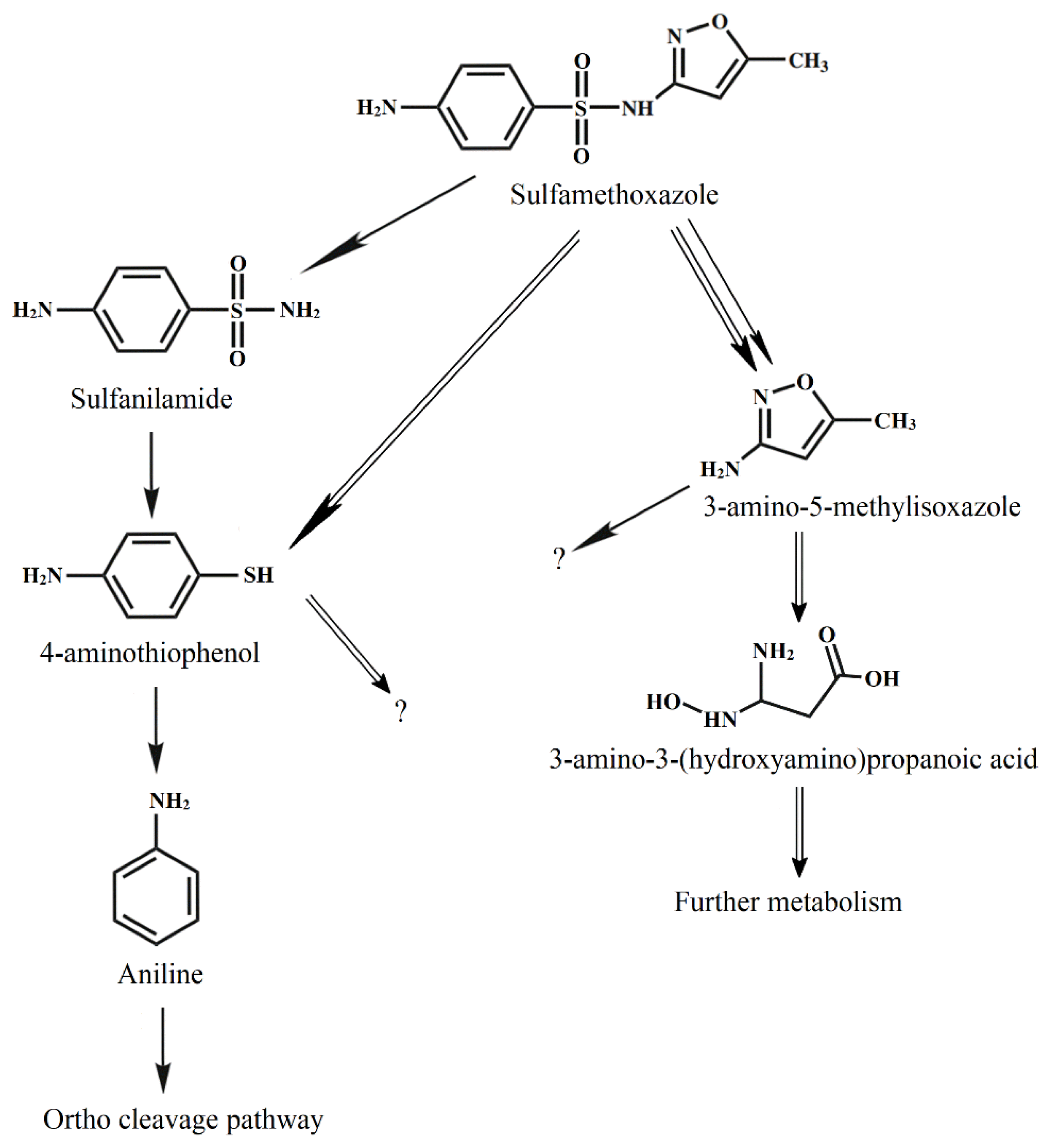
During the degradation of SMX by Acinetobacter sp. S1, some metabolites had m/z 93, 98, 125, and 172 (Fig. S2), indicating that aniline, 3-amino-5-methylisoxazole, 4-aminothiophenol, and sulfanilamide were produced, respectively [14]. The degradation of the substrate by Pseudomonas sp. S2 produced 4-aminothiophenol, 3-amino-5-methylisoxazole, and 3-amino-3-(hydroxyamino) propanoic acid (m/z 125). During the degradation of SMX by the mixed culture of Acinetobacter sp. S1 and Pseudomonas sp. S2, all these metabolites were also detected.
Acinetobacter sp. S1 first transformed SMX through hydrogenation and then degraded via desulfurization. This pathway was observed during substrate degradation by Pseudomonas psychrophila HA-4 [14] and Sphingobacterium mizutaii [31]. Acinetobacter sp. W1 transformed SMX into 4-hydroxyl-benzenesulfonic acid, N-hydroxy-acetyl-SMX, and the precursor of N-hydroxy-acetyl-SMX [17]. Under the anaerobic sulfate-reducing condition, SMX was transformed into five biodegradation intermediates, and most were altered in the isoxazole ring [35]. In another report, SMX could be converted into 56 intermediates under anaerobic conditions, whether further degradation of these intermediates was not known [36].
The determination of degradation of some metabolites showed that Pseudomonas sp. S2 exhibited a slow degradation of 4-aminothiophenol (25.7±5.8%) and sulfanilamide (14.5±3.4%) at 50 mg/L after 72 h, while data for 3-amino-5-methylisoxazole was 88.4±5.6%. In contrast, Acinetobacter sp. S1 degraded 77.4±8.8% and 18.2±5.1% of 4-aminothiophenol and 3-amino-5-methylisoxazole, respectively, at a chemical concentration of 50 mg/L. Moreover, Acinetobacter sp. S1 showed effective degradation of sulfanilamide (72.2±8.1%).
During the SMX degradation by Acinetobacter sp. S1, 4-aminothiophenol reached the maximum concentration at the 48th hour, and the concentrations then decreased (Fig. 6a). Meanwhile, the metabolite increased over time during the degradation by Pseudomonas sp. S2 (Fig. 6b). On the other hand, during the degradation of SMX by Pseudomonas sp. S2, 3-amino-5-methylisoxazole was produced, reached the maximum concentration at the 36th hour, and subsequently decreased (Fig. 6b). This product concentration increased with time during the degradation by Acinetobacter sp. S1. 3-amino-3-(hydroxyamino)propanoic acid was only produced during the degradation by Pseudomonas sp. S2 and its concentrations (< 1.0 mg/L) were consistently lower than those of 3-amino-5-methylisoxazole. 3-amino-3-(hydroxyamino)propanoic acid was entirely removed after 72 h. The sulfanilamide content did not exceed 1.2 mg/L during the degradation by Pseudomonas sp. S2 at 50 mg/L SMX.
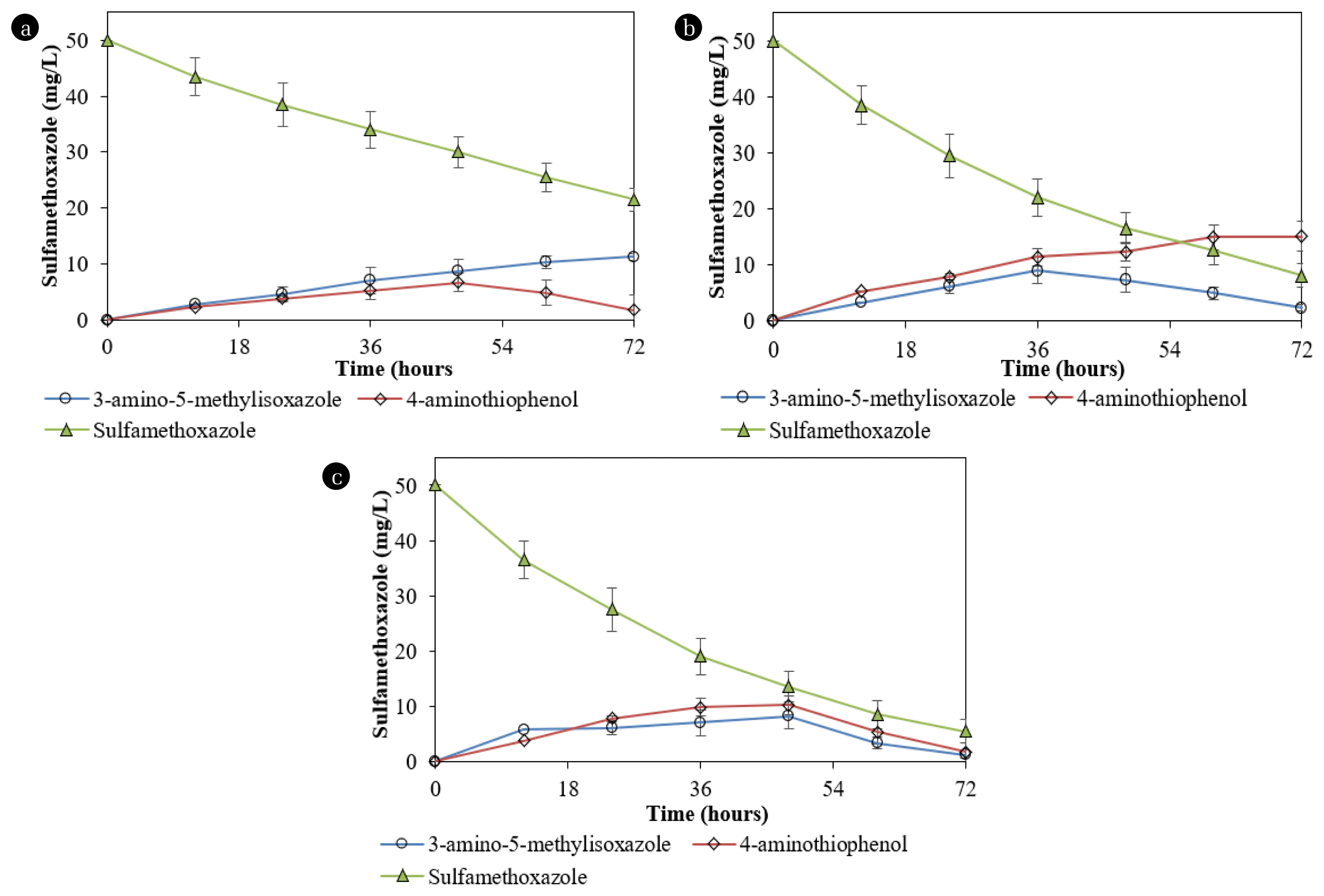
Degradation of SMX by (a) Acinetobacter sp. S1, (b) Pseudomonas sp. S2, and (c) mixed culture of Acinetobacter sp. S1 and Pseudomonas sp. S2 and production of degradation products.
SMX degradation by the mixed culture of both isolates was 89.0±5.5% for 72 h. During the SMX degradation by the mixed culture containing both strains, metabolites produced with the concentrations gradually increased until the 48th h and then decreased to not exceed 0.1 mg/L (Fig. 6c). The reductions in the concentrations of 3-amino-5-methylisoxazole and 4-aminothiophenol during the degradation by the mixed culture containing Acinetobacter sp. S1 and Pseudomonas sp. S2 indicated that each strain performed its role and cooperation. 3-amino-3-(hydroxyamino) propanoic acid was transformed from 3-amino-5-methylisoxazole by Pseudomonas sp. S2. The production of 3-amino-3-(hydroxyamino) propanoic acid was also recorded during the SMX degradation by Acinetobacter sp. [17].
The activities of enzymes extracted from the bacteria cultured on SMX and 4-aminothiophenol versus those of the cells cultured in the medium without these substrates are listed in Table 1. For the degradation by Acinetobacter sp. S1, the specific activities of catechol 1,2-dioxygenase were significant, whereas the activities of catechol-2,3-dioxygenase were negligible. The activities of catechol 1,2-dioxygenase extracted from Pseudomonas sp. S2 were significantly lower than those from Acinetobacter sp. S1. The results indicated that degradation occurred through ortho-cleavage pathways, and the aromatic ring cleavage was mainly conducted by Acinetobacter sp. S1. Based on the metabolites, the degradation pathways were proposed, as shown in Fig. 7.

Specific enzyme activities extracted from suspended cells grown on the MSM medium supplemented with ammonium sulfate and succinate.
4. Conclusions
The degradations of SMX by nonisolated microorganisms and isolated bacteria immobilized in PUF increased with the batch reactor operation using a pilot reactor, owing to increased immobilized biomass. The degradation rates at 10, 25 and 50 mg/L were 9.7±0.1 mg/day, 24.6±1.0 mg/day and 38.8±3.8 mg SMX/day under optimum conditions, respectively. Two SMX-degrading bacterial strains, Acinetobacter sp. S1 and Pseudomonas sp. S2 isolated from the immobilized biomass exhibited complete degradation of the antibiotic. During the degradation by the mixed culture of these isolated bacteria, aniline, 4-aminothiophenol, 3-amino-5-methylisoxazole, sulfanilamide, and 3-amino-3-(hydroxyamino) propanoic acid were produced as metabolites. The cooperation of both isolates in the mixed culture enhanced the degradation and reduced the concentration of produced metabolites. Therefore, this study demonstrates SMX degradation improvement using an immobilized biomass reactor and can serve as a reference for the degradation mechanism of the compound.
Supplementary Information
Acknowledgements
The author could like to thank all staffs in the laboratory of Dong Thap University who helped me do all experiments. I am grateful to the anonymous reviewers and editors whose suggestions helped to improve this manuscript.