Trends in Bioremediation of Heavy Metal Contaminations
Article information
Abstract
Heavy metal contamination of the ecosystem remains one of the severe global threats. Even in trace quantities, heavy metals and metalloids such as chromium, lead, mercury, cadmium, nickel, and cobalt are toxic and carcinogenic, posing a serious threat to human life. Certain microbes and plants have evolved detoxifying pathways to fight the harmful effects of these inorganic metals, paving the door for bioremediation. Because of its environmentally benign nature, economic viability, and low labor and effort requirements, bioremediation outperforms other approaches in eliminating heavy metals. This review highlights the potential of microbes on remediation of heavy metals in the context of environmental protection and also focuses on the critical tolerance mechanisms used by these microbes in combating heavy metal contaminations. Furthermore, the bioremediation potential of bacteria, fungus, algae, plants, biosurfactants, biofilms and genetically altered microorganisms for the removal of these heavy metals was reviewed in this study. Applying these techniques as a sustainable environmental technology in the near future has shown synergistic benefits with a many-fold increase in the removal of heavy metals.
Abstract
Graphical Abstract

1. Introduction
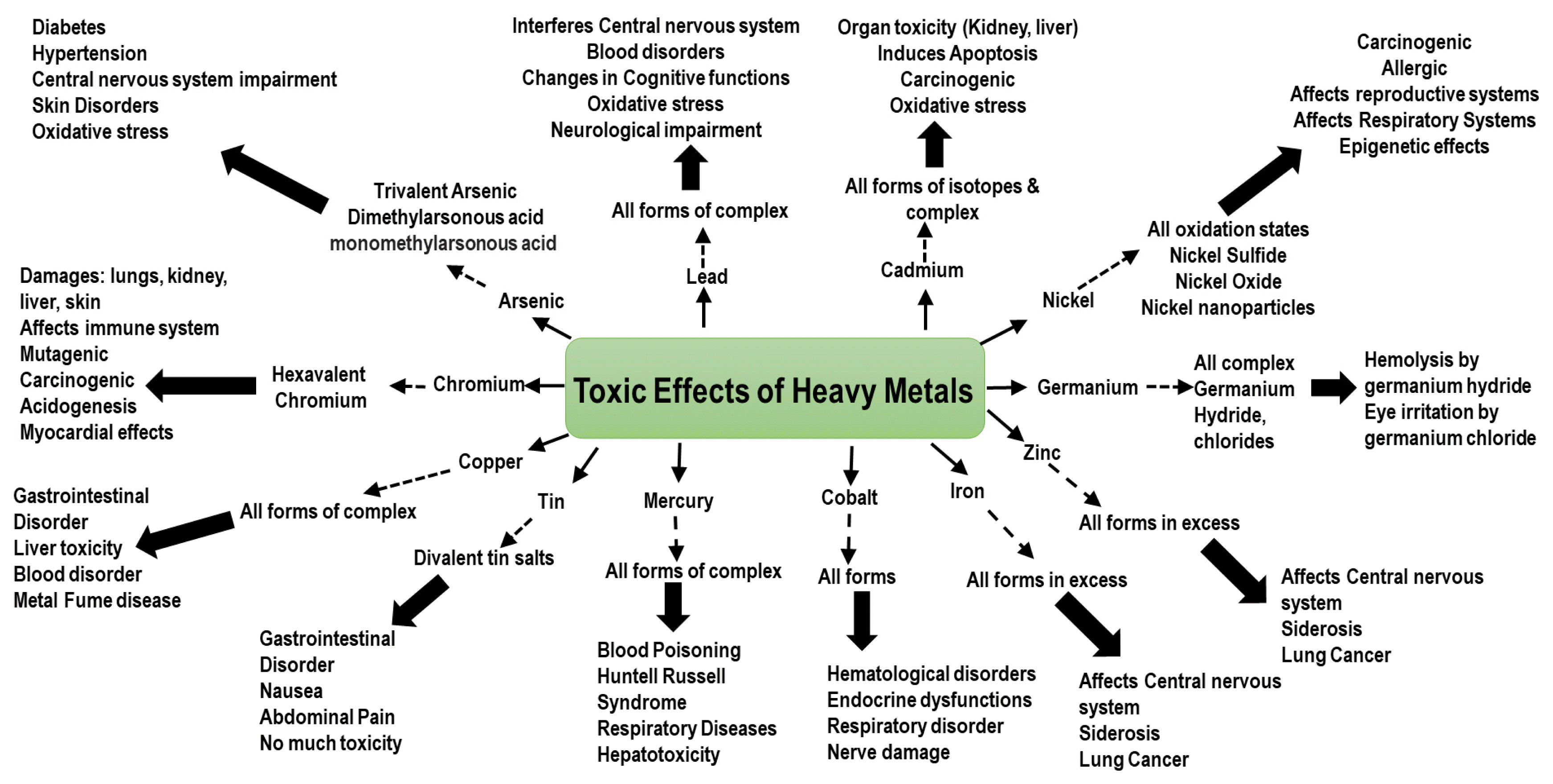
Rising heavy metal contaminations due to industrial growth pose substantial public health and environmental risk. Metals and metalloids having atomic weights and densities more than 50 and 4 g/cm3, respectively, are classified as heavy metals [1]. The typical heavy metals include silver (Ag), arsenic (As), nickel (Ni), zinc (Zn), cadmium (Cd), mercury (Hg), chromium (Cr), iron (Fe), lead (Pb), copper (Cu), vanadium (V), manganese (Mn), platinum (Pt), gold (Au), and aluminum (Al) [1, 2]. Other heavy metals include antimony (Sb), beryllium (Be), barium (Ba), bismuth (Bi), tin (Sn), germanium (Ge), gallium (Ga), indium (In), lithium (Li), molybdenum (Mo), selenium (Se), thallium (Tl), thorium (Th), and uranium (U) under different oxidation states [1]. Heavy metals are released into the environment in massive quantities from human activities and industrial operations, including fossil fuel burning, mining, electroplating, and wastes generated from textile industries, automobile industries, chemical industries, steel/iron industries, and petrochemical plants [3]. Furthermore, natural processes such as volcanic eruptions, ocean sprays, natural decay of organic matter, and wind erosions release heavy metals. According to US EPA standards, the maximum allowable concentration for Pb, As, Hg, Cd, and Cr are 0.015, 0.01, 0.002, 0.005, and 0.1 mg/L, respectively [4]. Heavy metals tend to possess a high biological half-life (lead has a half-life of 5000 years) credited to their non-biodegradable nature, causing bio-accumulation and biomagnification in the food chain leading to serious environmental and health consequences [5]. Direct contact, ingestion, or inhalation of these heavy metals, even at low concentrations, is often poisonous, affecting the central nervous system with mutations and genetic damage, increasing the risk of cancer and teratogenic effects [6, 7]. Fig. 1 represents the various metals and their possible toxicity to humans and the environment. As a result, before releasing wastewater effluents, it is critical to remediate these harmful heavy metals.
Several technologies are being implemented to remove heavy metal ions, broadly classified as physico-chemical and biological methods [8]. The conventional physico-chemical methods include solvent extraction, evaporation, chemical oxidation, ion exchange, reverse osmosis, electrochemical methods, precipitation, ultrafiltration, nanofiltration, leaching, land filling, magnetic separation, and other mechanical separations [8]. Furthermore, these methods consume less time with high controllability and pliability to higher concentrations of heavy metals. However, most of these approaches are prohibitively costly, inoperable at low heavy metal concentrations below 100 mg/L, and releases hazardous derivatives/solvents in high quantities that are harmful to the environment [6, 7].
As an alternate, a green technology utilizing several biological agents, primarily bacteria, algae, yeasts, molds, and plants, has garnered increased attention for heavy metal removal and recovery [9]. Unlike physico-chemical methods, biological agents add scope for economic feasibility attributed to their high heavy metal removal efficiencies, cost-effective nature, and ease of availability. The utilization of biological agents for removing toxic compounds is termed as bioremediation. In the case of heavy metal removal, the bioremediation process targets converting high valent toxic heavy metals into less toxic ions. These biological agents have numerous binding sites with a large surface area to volume ratio and strong binding affinity that offers high removal efficiency for heavy metals [10]. These biological agents also exhibit exceptional natural properties such as high photosynthetic efficiency (restricted to photosynthetic microbes and plants) and a basic structure to thrive in harsh environments such as heavy metal contamination, high salinity, nutritional stress, and severe temperature. The properties of biological agents (i.e., living or non-living), the characteristics of the targeted metal, the type of binding sites involved in metal sequestration, operational parameters (temperature, pH, contact time, concentrations of metal ions and biological agents), and the characteristics of metal solution and the presence of competing co-ions are few factors that govern the efficiency of bioremediation mechanisms. These intrinsic features allow the use of biological agents in soil or water remediation [9].
Furthermore, the bioremediation process generates biomass waste that can be redirected to agricultural industries as nutrient sources for plants’ growth, making them exclusive. However, the current state-of-the-art technologies suffer significantly in terms of toxic intermediate formation, process sustainability, and economic feasibility [11, 12]. Over the decade, extensive research has been conducted to address these issues with novel genetic engineering approaches and process strategies [3–5] and are updated at high rates. Therefore, this review focuses on the trends in heavy metal bioremediation, mechanisms involved, biological agents used, with a clear understanding of the prevailing hurdles and prospects in a consolidated manner.
2. Mechanisms of Bioremediation
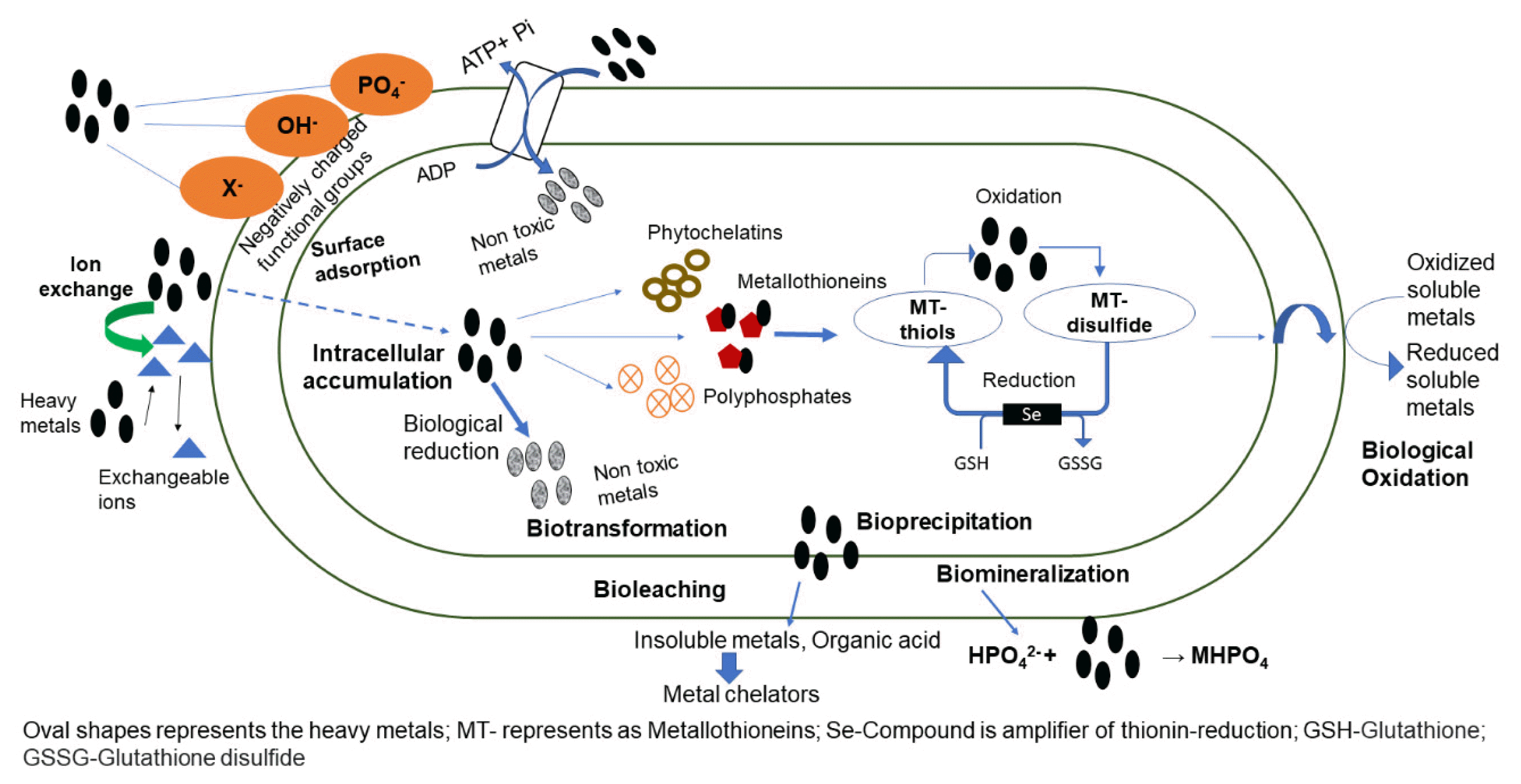
Even though heavy metals are typically poisonous, organisms have evolved specific resistance mechanisms and complex intracellular pathways to utilize and detoxify heavy metals for cellular reproduction [13]. For instance, few bacteria and cyanobacteria have developed a mechanism to synthesize extracellular polysaccharides (EPS), which readily binds to heavy metals and avoid the entry into the cells. In other bacterial, fungi, and algal cells, the heavy metals readily cross the extracellular membrane and accumulate in the cytoplasm, where they get detoxified with intrinsic metabolic reactions or expelled out of the cellular structures through efflux pumps [14]. These mechanisms are highly influenced by the nature of the organism and its essential proficiencies. Based on these capabilities, the bioremediation of heavy metals is achieved through several mechanisms, which are not limited to biosorption, bioaccumulation, bioreduction, bioprecipitation, biovolatilization, bioleaching, composting, land farming, bioreactors, biopiles, and biosparging [15]. Phytostimulation, and rhizofiltration are the other two mechanisms that can be evidenced in case of phytoremediation [15]. Among them, biosorption and bioaccumulation are two different mechanisms that are profound across various organisms, including plants.
The biosorption mechanism is a complicated process in which biosorbents interacts and adsorbs the metal ions in the surrounding environment. As a biosorbent, several natural resources are exploited, such as biomass of bacteria, fungi, algae, agricultural wastes, plants, and industrial by-products. Usually, the dead biomass or cells are used for biosorption, which can remove heavy metal contaminants in extremely low concentrations ranging in parts per billion [16] with high efficiency. In some instances, live cellular biomass is actively utilized to remove heavy metals via biosorption [16]. Irrespective of whether it is a dead or live biomass, biosorption is a surface phenomenon that allows the adsorption of heavy metals to the cellular surface. The mechanisms involve surface adsorption due to physical interaction (electrostatic or Van der Waals interaction), chemical interaction (ion exchange displacement of attached metal cations), complexation, diffusion, or precipitation [17]. The cellular surface usually bears a negative charge that strongly attracts the heavy metals, whereas, in some instances, this cellular surface may comprise a mucus or a polysaccharide layer that adsorbs the heavy metals strongly through physical interactions. Heavy metals are adsorbed on the surface passively without requiring energy expenditure until equilibrium is reached [18]. Cell surfaces comprise functional groups such as phosphates, sulfates, amides, hydroxyl, or negatively charged proteins that exchange ions with the metal ion resulting in strong ionic interaction. In the case of live cells, the interaction is usually the same; however, the metal ions may reach the periplasmic space through porins available on the surface and may establish interaction with the cellular membranes. The ionic interactions found with heavy metals allows the reduction of heavy metals and its detoxification. The aggregation of two or more metal species and functional groups on the cell surface is termed as complexation. Monodentate and polydentate complexes are the two forms of complexes predominantly found in biosorption [19]. The metal ion forms covalent connections in the center with the ligands in monodentate complexes, whereas multiple metal ions bind to the ligands of polydentate complexes at numerous sites. Using living biomass for sorption may not be viable due to toxic metals that may accumulate in cells and impede metabolic function, resulting in cell death. Dead biomass, on the other hand, is unaffected by toxicity, requires no growth/nutrition medium, and is adaptable to changing environmental circumstances.
The absorption of contaminants by living biomass/cells is known as bioaccumulation, and it includes metabolism dependent active uptake of heavy metals. The metabolism-dependent trafficking mechanisms involving passive diffusion, ion pumps, protein channels, and carrier-mediated transport determine the heavy metal removal efficiency from the surroundings [18]. The heavy metal transport across the phospholipid bilayers of living cells occurs in two phases. The adherence of heavy metals to the cell surface in a metabolism independent fashion is the initial step, followed by the internalization of metal ions via the cell membrane in the second step [16]. This resembles the cellular uptake of essential ions like Na2+, K+, and Ca2+. The metal cations with equal charge and ionic radius hijack the essential ion channels and enters the intracellular phase [16]. Active transporters are evidenced for cadmium, chromium, silver, lead, mercury, strontium, uranium, thorium in Pseudomonas putida, Bacillus subtilis, Thiobacillus ferrooxidans, Rhizopus arrhizus, Micrococcus luteus, Saccharomyces cerevisiae, and Aspergillus niger, respectively [20]. The intracellular reduction of heavy metals follows a variety of metabolic pathways.
Bioprecipitation refers to microbial activities that transform soluble metal species and metalloids into insoluble forms such as hydroxides, carbonates, phosphates, and sulfides [21]. The metal precipitates formed remains attached to the microbial cells, and slight pH changes result in easy recovery of heavy metal ions. In general, the microbes release organic acids, electron donors into the extracellular medium, which precipitates the heavy metal ions. In some cases, enzymes precipitate the heavy metal ions. For instance, alkaline phosphatase derived from calf intestine were found to precipitate cadmium, nickel, cobalt and chromium ions obtained from the tannery and electroplating industries [21]. Unlike bioaccumulation, bioprecipitation is not always a metabolism-dependent process, rather the environmental changes induced by the microbes play a key role in the precipitation. For instance, changes in the pH values, and redox potentials in the extracellular environment augments the precipitation of heavy metals and its stability. Pseudomonas aeruginosa and Klebsiella planticola are known to induce precipitation of cadmium ions [21]. Under anaerobic circumstances, sulfate-reducing bacteria accumulates hydrogen sulfide derived from organic substrates, that precipitates metal ions in sediments as insoluble metal sulfides (e.g., CuS, PbS, ZnS, and SeS) [22, 23]. As compared to hydroxides and other precipitates, sulfide-based precipitation allows near complete removal of heavy metal ions effectively in the presence of both organic and inorganic electron donors. Biological oxidation of soluble ferrous ion, forms amorphous iron(III) hydroxides or crystalline iron oxides, which coprecipitate other metals like As, Cd, and U indirectly [24]. These insoluble metal compounds considerably reduce metals’ leaching potential and bioavailability in sediments. However, because biological oxidation of sulfide minerals or biological reduction of iron(III) oxides might resolubilize the solidified metals back into the aqueous phase, it is critical to monitor changes in ambient redox potential and induced microbial activity. Other iron precipitates available are scorodite, schwertmannite, ferric hydroxide, geothite, hematite, and jarosite.
Biomining is a broad word that encompasses both bioleaching and bio-oxidation. The complexation and dissolution of heavy metals from sediment ores by the organic acids produced by acidophilic microbes is termed as bioleaching. For instance, Streptomyces albidoflavus an actinomycete, has been reported to leach out different metal ions with varying efficiency from printed circuit board [25]. The study showed a maximum removal of zinc up to 82%, followed by nickel (81%), calcium (74%), copper (68%), aluminum (66%), cadmium (65%), silver (56%), lead (46%), and iron (42%). Such bioleaching methods are usually applied to remove heavy metals from solid particles or soil.
Microorganisms can interact with heavy metals and influence their biotransformation by oxidation, reduction, methylation, demethylation, and complexation [26–28]. The effects of microbial transformation on heavy metal accumulation and transformation are determined by the physico-chemical properties of metal forms as well as the geochemical circumstances of polluted locations [29]. Mineral components usually comprise significant levels of insoluble metals that are not biodegradable. In such cases, microbial mobilization can be used to regulate the release and subsequent transformation of insoluble metal species. In case of soluble metal species, microbes driven transformation is essential for modifying the speciation by modulating the oxidation/reduction states of heavy metals in sediments, solubility, mobility, bioavailability, and toxic nature. The bioreduction of Cr(VI) has been linked to a combination of biological processes, including direct reduction of Cr(VI) to Cr(III) and indirect reduction of Cr(VI) to Cr(III) by bio-produced Fe(II) [30].
In many microbes, the intracellular heavy metals bind to metallothionein complex, which involve in a sequence of redox reactions that results in the reduction of metal ions [31]. The metallothionein is a cysteine-rich protein predominantly found in cyanobacteria, higher microbes and organisms with high affinity towards metals such as zinc, cadmium, copper, and mercury. The genes for the synthesis of metallothionein are also conserved in the genus Pseudomonas, which provides additional benefits for these organisms to survive at toxic environmental conditions [32]. The redox potentials of metallothionein complexes directly affects the cellular super-oxide radicals and hydroxyl radicals [31]. Fig. 2 depicts the various mechanisms in different organisms that allows to tolerate and reduce heavy metals.
2.1. Bacterial Bioremediation Mechanisms
As compared to other microbial forms, bacteria are well known for their robustness, large surface area availability, faster growth rate, and high resistance towards toxic heavy metals [33] making them unique. The bacterial strains have evolved several pathways for managing heavy metals, which includes efflux pumps, redox reactions, complex formations, extra and intra-cellular sequestrations, and in certain cases the metal ions are used as electron acceptors in oxidative phosphorylation. For example, heavy metal protein complex formation and reductive precipitation were the two resistance mechanisms depicted by Clostridium spp. [34]. Further, the resistance towards these heavy metals occurs either in their chromosomal genome or in the plasmid genome. For instance, the resistance genes for zinc, and antimony were reported in bacterial plasmid genome that usually code for efflux transporters [33]. The other heavy metal resistance genes against arsenic, copper, and cadmium were found abundant in the genomic DNA of Proteobacteria and Actinobacteria [34]. Several strains of bacteria such as Yersinia enterocolitica, Geobacter sp., and Deferribacter desulfuricans are shown to possess ars operon in their genome, which is regulated in response to the extracellular arsenic concentrations. The ars operon comprises a gene acr3 that encodes an arsenic efflux protein, which supports that efflux is one of the prominent resistance mechanisms available in bacterial systems.
The presence of extracellular polymers and charged functional groups forms better complex with heavy metals and reduces the concentration in the surrounding environment. There are variety of extracellular polymers of definite negative charge that attracts the heavy metals. Analysis of EPS from the non-living biomass of Micrococcus luteus and Kocuria palustris using infra-red spectroscopy showed the presence of amino and phosphate groups that contribute to Cd binding and complex formation [35]. Bacillus sp. S3 is an unique strain with multi-metal tolerance against antimony, copper, hexavalent chromium, and cadmium attributed to the thick EPS layers available on its surface [36]. A significant increase in the concentration of extracellular polysaccharide is evidenced when exposed to cadmium, chromium and copper ions resulting in detoxification [36]. Azotobacter bd39 is also known to alleviate mercury toxicity with EPS and offers resistance to the host plant [37]. Xanthan, alginate, hyaluronic acid, gellan, and fucopol are certain anionic polymers present on the surface of different organisms such as Xanthomonas campestris, Pseudomonas aeruginosa, Streptococcus sp., Sphingomona spaucimobilis, and Enterobacter sp., respectively [38]. An exhaustive list of different exopolysaccharides and associated role in heavy metal removal are reported [37].
Metal binding through precipitation is caused by metal ions reacting with functional groups on the bacterial surface, resulting in insoluble organic metal precipitates that stay connected to the microbial cells. This ion exchange process involves the exchange of binary metal cations with a counter ion on the biosorbent’s surface. For Pb(II) adsorption by Klebsiella sp. J1 an ion exchange mechanism involving the counter ions K+ and Mg2+ was discovered in the presence of functionalized extracellular polymer [39]. Bacillus laterosporus and Bacillus licheniformis were used to successfully biosorb Cd and Cr ions from metal solutions with the same mechanism [16]. Some harmful heavy metals such as Ni(II), Cd(II), Cr(III), Cr(VI), and Co(II) were removed from diverse industrial effluents by Escherichia coli C90, which generates alkaline phosphatase to precipitate the metal cations [16]. Evaluation of dead and living forms of the strain 1369SI of Bacillus salmalaya for chromium removal showed varied efficiencies and removal rates. The maximum adsorption capacity for the dead biomass was found to be 20.35 mg/g, which was ~57% higher than adsorption capacity of living biomass [40]. The cadmium tolerance levels of actinomycetes were far better than that of gram −ve strains and gram +ve strains. Similar result was observed in removal of lead, depicting maximum removal by Azotobacter strain up to 37%, which was ~4 folds higher than M. luteus strain [41]. However, thick peptidoglycan layer and negative surface charge availability in gram +ve strains increases the binding of metal cations as compared to gram −ve strains [33].
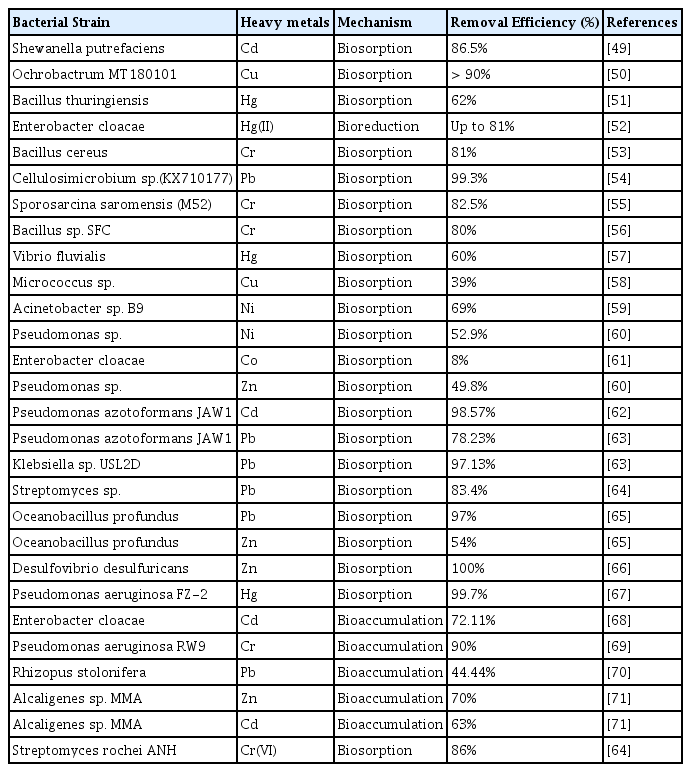
Under different environmental circumstances, such as pH (7–9), temperature (30–40°C), and Cr(VI) concentrations (50–250 mg/L), a Bacillus isolate was able to physiologically convert Cr(VI) to Cr(III), improving Cr removal efficiency [42, 43]. At low and high pH, functional groups such as amine, carboxylic, phosphate, and hydroxyl are protonated and deprotonated, respectively. Under low pH conditions, the excess H+ ions compete with metal ions, whereas at high pH levels metal removal is reduced because of repulsive interactions between metal anions and negatively charged functional groups [41]. Under the ideal circumstances of pH 7 and 37°C, the Bacillus isolate totally reduced Cr(VI) at a concentration of 120 mg/L in 48 hours [43]. Using acetate as a carbon source, a mixed anaerobic culture of Anaerolineaceae, Spirochaeta, and Spirochaetaceae has the ability to bioreduce V(V) to V(IV) and Cr(VI) to Cr(III), with removal efficiencies of 97.0 and 99.1 percent for V(V) and Cr(VI), respectively [43]. In the presence of hematite and dissolved organic matter, Geobacter sulfurreducens exhibited the ability to remove Cr(VI). The anaerobic sulfate-reducing bacteria are known for its inherent capabilities in sulfate and heavy metal removal. The sulfides synthesized by these bacteria utilizing organic acids and hydrogen as substrates, reacts and removes heavy metals as metal sulfides [45]. In the presence of zero valent iron, the toxicity exerted by heavy metals on sulfate-reducing bacteria is significantly reduced, while assisting in establishment of anaerobic condition by reducing redox potential [45]. For instance, in the presence of zero valent iron, the sulfate-reducing bacteria demonstrated 99% removal of hexavalent chromium and divalent zinc, with limitations in divalent manganese removal [45]. The other type is the magnetotactic bacteria falls under the family alphaproteobacterial, deltaproteobacteria, gammaproteobacteria, and nitrospirae [46]. The unique future of magnetotactic bacteria is its magnetotaxis nature that allows alignment of bacterium according to applied magnetic field attributed to the presence of magnetosomes made up of greigite and magnetite. This property adds scope for easy recovery of used bacterium and removed metal ions. These magnetotactic bacteria also enriched with extracellular charged functional groups allowing substantial removal of metal ions such as silver, chromium, plutonium, copper, aluminum, and cobalt [46]. The key advantage of using magnetotactic bacteria is the time invested in the removal of heavy metals. For instance, within 10 minutes up to 77% removal of Cr(VI) by magnetotactic culture was reported in the presence of electric field, supplemented with cobalt and copper ions at pH 6 [47]. Another study showed a maximum removal of Co up to 1.16 g/g dry weight by the magnetotactic bacteria Alphaproteobacterium MTB-KTN90 at optimal operational conditions [48]. Several bacterial strains in both live and dead forms are known to have bioremediation potentials against heavy metals, which are enlisted in Table 1.
2.2. Fungal Bioremediation Mechanisms
Fungi are well-known for their ubiquitous presence in nature and are often used in industrial applications [72]. Fungi are suited to their environment (in terms of morphology, ecology, and metabolism) and are responsible for activities such as decomposition and nutrient cycling under natural settings [73]. The fungal strains are also robust in nature in terms of resisting harsh environmental conditions such as high moisture, nutrient conditions, and pH. Mycoremediation is the use of fungus (either alive or dead) to remove toxins from various environmental segments [74, 75]. Mycoremediation is a low-cost method that produces no hazardous waste. As a result of the complete mineralization of the contaminants in nature, it provides a comprehensive solution [21]. The selection and use of an appropriate fungal species for the target heavy metal or other pollutants is critical to the effectiveness of mycoremediation. Fungi have the capacity to efficiently store heavy metals in their fruit bodies, rendering them inaccessible or lowering their concentration in the media [21]. The future availability of heavy metals and other pollutants in the medium is determined by the life of the fungus, the chemical behavior of the elements, and the presence or absence of the fungi following sequestration. Since heavy metals may cause cell lysis and death in fungi, multiple research have been conducted to determine the extent to which different fungi can tolerate them. At the genus and species levels, it was deduced that specificity and maximum tolerance limit for metals differ substantially. Among the most resistant strains are Aspergillus sp., Penicillium sp., and Fusarium sp. [76]. For example, Aspergillus flavus (ASC1) and Aspergillus niger (ASB3) have shown to grow in an medium enriched with 1000 ppm of As, with subsequent removal of As from the medium [76]. Aspergillus sp. and Fusarium sp., have depicted high resistance against Pb, and Cr, and slightly reduced resistance against Cu, and Zn [77]. A. flavus showed tolerance up to 0.4 g L−1 whereas other strains of Penicillium sp., and Fusarium sp., showed tolerance up to 0.1 g L−1 [77]. Another study depicted the capability of Aspergillus sp. to extract 65% of Cr from tannery effluent compared to 85 percent from the synthetic medium [78]. Cr has been observed to be tolerated by Penicillium chrysogenum and Trichoderma viride up to a concentration of 600 mg L−1 [79]. Similarly, a study of 36 Anthrodia vaillantii strains revealed that some were able to survive up to 40 mM of Cu.
Saccharomyces cerevisiae has been shown to sequester up to 65–79 percent of Pb and Cd from contaminated soil [80]. The biosorption process includes fungal cell walls (which contain chitin, proteins, glucans, lipids, pigments, and polysaccharides) and functional groups such as hydroxyl, carboxyl, amino, sulphate, or phosphate, and is mediated by interactions such as adsorption, ion exchange, and complexation [81, 82].
Wood-decaying species (white- and brown-rot fungus), mushrooms, and other fungi all employed for mycoremediation because of their ability to absorb heavy metals in their fruiting bodies. Contact time, mycelia age, and fructification influences the metal absorption in mushrooms. Heavy metals can accumulate at high levels in some edible wild mushroom types. Pleurotus ostreatus and Termitomyces clypeatus are two species of white-rot fungus that have been shown to digest persistent pollutants such as heavy metals [83]. In many cases, exposure of fungal strains to the heavy metal contaminated soil or wastewater induces the expression of resistance genes associated with remediation of heavy metals. For example, Suillus luteus from metal-contaminated locations were shown to be more resistant than those from non-contaminated places. However, varied remediation scenarios are observed when a Cd-resistant fungus, P. betulinus, isolated from both polluted and non-contaminated sites depicted no correlation. According to the foregoing findings, there is a significant variation in metal tolerance capacity and specificity. This is most likely owing to the application of various techniques to combat metal toxicity. The capacity of metal tolerance in fungus is also influenced by soil physiochemical characteristics and carbon content [74, 77, 84]. The recent trends in utilization of fungal biomass for bioremediation of heavy metals are provided in Table 2.
2.3. Algal Bioremediation Mechanisms
Heavy metals including boron, cobalt, copper, iron, molybdenum, manganese, and zinc are used by microalgae as trace elements for enzymatic processes and cellular metabolism, however other heavy metals like As, Cd, Cr, Pb, and Hg are poisonous to microalgae. Low hazardous heavy metal concentrations augment growth and metabolism of microalgae due to the hormesis phenomena [90]. Because of their resilience to heavy metal stress, several cyanobacterial species, such as Anabaena, Oscillatoria, Phormidium, and Spirogyra, naturally thrives in heavy metal-contaminated water [91]. Microalgae have reactive groups with active binding sites that can form complexes with contaminants in wastewater, in addition to heavy metals. This causes flocculation, which decreases the total dissolved solids and total suspended solids concentration [6].
A two-stage mechanism is used by microalgae to remove heavy metals. The rapid extracellular passive adsorption is the first stage, while the slow intracellular positive diffusion and accumulation is the second. The cell wall of microalgae is primarily composed of polysaccharides (cellulose and alginate), lipids, and organic proteins, and provides many functional groups (such as amino, carboxyl, hydroxyl, imidazole, phosphate, sulfonate, thiol, and others) capable of binding heavy metals in addition to cell polymeric substances such as peptides and EPS with uronic groups [92]. They also possess a lot of monomeric alcohols, laminaran, deprotonated sulfate, and carboxyl groups, which attract both anionic and cationic heavy metal species. These variety of functional groups and EPS in microalgae biomass contribute to efficient biosorption of heavy metals [93]. Transcriptomic analysis of several microalgal strains revealed the increased expression of transporter genes related to the uptake of specific heavy metal ions and divalent cations [94] in the presence of extracellular heavy metal ions. For instance, Auxeochlorella protothecoides showed higher expression levels of natural resistance-associated macrophage proteins in the presence of increased cadmium concentration in the extracellular medium [94]. The other specific transporters available are Zrt, irt-like transporters, iron transporter, P-type ATPases, copper transporter, arsenite efflux transporter, multidrug and toxic compound extrusion protein, and ATP-binding cassette transporters [94].
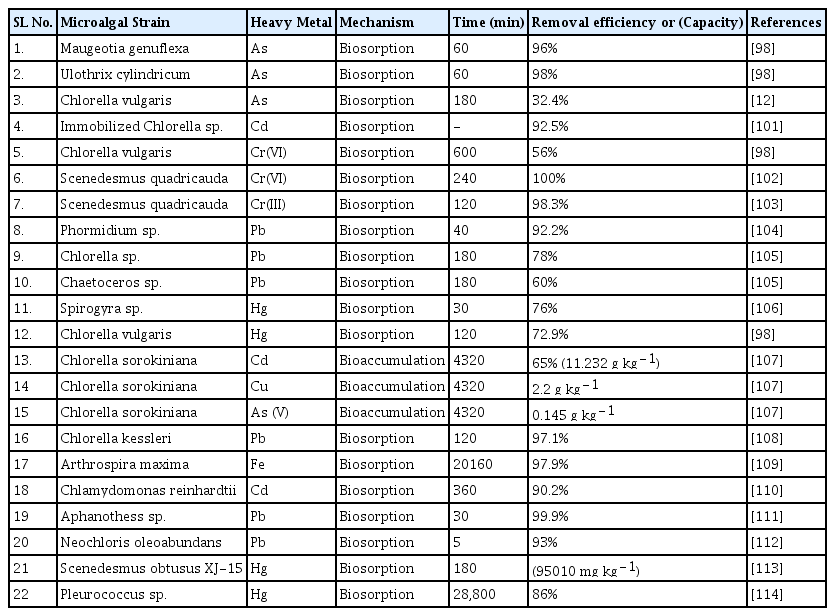
Microalgal species employ heavy metal immobilization, gene regulation, exclusion, and chelation, as well as antioxidants or reducing enzymes that reduce heavy metals via redox processes, to defend themselves against heavy metal toxicity [95]. Microalgae creates protein-heavy metal complexes in their cells without affecting their own function [94]. The organometallic complexes are further segregated inside vacuoles to manage the homeostasis of heavy metal ions in the cytoplasm, while reducing the hazardous effects [96]. Heavy metals also stimulate the production of phytochelatins (PCs), which are thiol-rich peptides that interact with heavy metals to reduce their effects [95]. Microalgae produce antioxidant enzymes such as ascorbate peroxidase, catalase, glutathione reductase, peroxidase, and superoxide dismutase (SOD) to combat free radicals released by heavy metals during adsorption, as well as non-enzymatic antioxidants such as carotenoids, cysteine, ascorbic acid, glutathione and proline [97]. SOD serves as the first line of defense against the increased superoxide anions released in the cells in response to heavy metal concentrations. Catalase further degrades hydrogen peroxide into water and oxygen molecules [96]. Cysteine acts as an indication for the production of several antioxidants by acting as an indirect or direct precursor for PCs, glutathione, metallothioneins, and other sulfur-containing molecules. Ascorbic acid and glutathione are essential endogenous antioxidants produced by microalgae that play a vital role in the reduction of reactive oxygen species (O−, ROS) and free radicals [98]. Aside from maintaining the balance of O− generation and removal, ascorbic acid protects microalgal cells by regulating the ascorbic acid-glutathione pathway and the activity of metal-containing enzymes, as well as the dissipation of excess excitation energy. Furthermore, microalgae release a high quantity of ascorbic acid as a hydrophilic redox buffer that protects the cytosol and other cellular components from oxidative hazards [93, 99]. High levels of glutathione, on the other hand, protect microalgae by providing tolerance, scavenging free radicals, accelerating PCs and ascorbic acid production, and restoring substrate for other antioxidants [95, 97]. Addition of phytohormones are known to augment the response of algal cells against heavy metal stress by detoxifying reactive oxygen species. For example, absicisc acid and brassinosteroids are known to accumulate in the microalgal strains Chlorella vulgaris, and Acutodesmus obliquus exposed to high levels of lead, copper, and cadmium resulting in PCs activation [100]. Table 3 represents the removal efficiencies obtained for different algal strains.
2.4. Plant-assisted Remediation of Heavy Metals
Plants and their associated rhizosphere microorganisms are used in phytoremediation to clean up heavy metal-contaminated sediments. The accumulation of heavy metals in plant system involve a series of steps, which includes mobilization of heavy metals for effective adsorption, uptake by root systems, loading of xylem, root to shoot transport, compartmentation, sequestration, and detoxification [115]. The mobilization of heavy metal ions occurs through the exudates synthesized by the plants and microbiome in the rhizosphere. Plant roots uptake the heavy metal ions through active and passive transport systems, which further forms complexes with organic acids such as carbonates, phosphate, sulfates and are immobilized in cellular walls or in intracellular vacuoles. The stored heavy metal ion complexes in vacuoles are transported in to xylem and shoots [115]. Phytoremediation of heavy metals may be divided into four categories based on the major processes in the plant metal system: phytostabilization, phytoextraction, phytofiltration, and phytotransformation. Among these processes, phytoextraction has been widely used as a remediation technology, which generally involves three steps: the cultivation of suitable metal accumulating plants, the harvest of metal-enriched plant biomass, and post-harvest treatment of plant biomass for added market value (e.g., energy recovery from thermal treatment).
Many variables influence phytoextraction efficiency, including metal speciation and bioavailability, soil characteristics and plant species, and rhizosphere microorganisms [80]. Except for hyper-accumulating plants, direct absorption and accumulation of heavy metals by plants is typically negligible. In such cases, the microbial population of the rhizosphere is critical for heavy metal bioremediation. Stimulation of microbial activity, microbial redox transformation, and microbial catalyzed precipitation of insoluble metal compounds in the rhizosphere are some of the tactics that improves the phytoremediation efficacy of heavy metal–contaminated sediments. As a result, more emphasis is being directed to the use of diverse microorganisms/microbial processes such as bioleaching, biosorption, bioaccumulation, bioprecipitation, and biotransformation to remediate heavy metal polluted sediments [91].
2.5. Removal of Heavy Metals by Biosurfactant, and Biofilms
2.5.1. Heavy metals removal by biosurfactant
Biosurfactants are amphiphilic surface-active chemicals generated by a range of microorganisms such as bacteria and fungus have garnered increasing interest as an alternative to chemical leaching agents such as synthetic surfactants. The biosurfactants have low surface and interfacial activity, low toxicity, high biodegradability, high pH and temperature tolerance, potential environmental compatibility over synthetic chemical surfactants [116]. With wide variety of chemical structures biosurfactants show wide range of binding capability with different metal ions. Numerous surfactants were obtained from different microbes such as Burkholderia sp., Bacillus spp., Pseudomonas spp., Candida spp., and Citrobacter freundii [117]. Burkholderia sp. Z-90 developed a biosurfactant glycolipid that was employed as a bioleaching agent to remove mixed hazardous metals such as Zn, Pb, Mn, Cd, Cu, and As from polluted soil. Because of their large acid-soluble proportion and greater complexation with biosurfactant, Mn, Zn, and Cd were more successfully removed from soil [118]. The increased metal-biosurfactant complexation of the biosurfactant generated by Pseudomonas sp. CQ2, depicted a better removal efficiency than conventional chemical leaching agents (i.e., SDS and Tween-80) for the bioleaching removal of Cd, Cu, and Pb [119]. Furthermore, heavy metals were effectively removed from polluted river sediment using the biosurfactant rhamnolipid in exchangeable, carbonate bound, or Fe-Mn oxide–bound fractions [120]. An anionic lipopeptide obtained from marine sponge associated Bacillus sp., showed high removal efficiency of mercury, manganese, lead, and cadmium up to 75.5%, 89.5%, 97.73%, and 99.93% respectively at 2× critical micelle concentration [121]. However, due to the poor manufacturing yield of biosurfactants and complexities in purification, the wide use of biosurfactant-induced technologies are currently limited. To improve biosurfactant production and broaden biosurfactant uses, future work should focus on improving identification of novel biosurfactant producers, fermentation operations (i.e., inoculum, pH, temperature, and mixture conditions), medium feeding (i.e., carbon and nitrogen sources), genetic engineering, and design of cost-efficient downstream process. Further, commercial scale experimentation should be opted over laboratory scale results in order to realize the potentials of biosurfactants on economical heavy metal removal [117].
2.5.2. Removal of heavy metals by biofilms
A biofilm is a collection of microbial cells adhered to a surface and encased in a matrix of extracellular polymeric molecules. Bacterial biofilms are aggressive than free living bacteria and resistant to adverse circumstances such as stress conditions (pH fluctuations, temperature changes, nutrient availability) by enabling coordinated sensing and response elements in terms of quorum sensing [122]. Biofilm has a high sorption capacity, high tolerance for hazardous inorganic components, even at deadly doses and a low production cost; hence it is gaining popularity in wastewater treatment. Metal removal effectiveness ranged from 4.79 percent to 10.25 percent for planktonic cells and from 91.71 percent to 95.39 percent for biofilm cells, according to research on Rhodotorula mucilaginosa [115]. In certain cases, the presence of heavy metal ions in the extracellular conditions induce biofilm production. For instance, in the presence of Cr(VI), magnesium, and nickel, Arthrobacter sp., SUK1205 produced a strong viscous biofilm, which also activated the removal of these metal ions effectively [123]. The biofilm of Arthrobacter sp. SUK1205 showed a complete removal of 0.5 mM Cr(VI) within 4 days of treatment [123]. A similar study showed the performance of the biofilm of Arthrobacter sp., on the removal of Cr(VI) in a packed bed reactor under batch and fed-batch operational modes. The fed-batch studies showed a chromium removal rate of 0.49 mg L−1 h−1 under industrial model solutions, whereas the rate was 0.79 mg L−1 h−1 when operated under laboratory conditions [123].
The use of an E. coli ASU 7 biofilm supported on granulated activated carbon improved Cr(VI) biosorption compared to the bacterium’s lyophilized biomass and the solitary granulated activated carbon [124]. At lower concentrations of Pb(II) and Fe(II), E. coli ASU 7 biofilm was employed to remove both the metal ions with 100% effectiveness. Fixed bacterial cells had a greater biosorption efficacy than suspended free cells for the removal of Pb(II) and Cu(II) [125].
Bacillus circulans biofilm was able to absorb 78 percent and 40 percent of Pb(II) and Cr(VI), respectively from an initial concentration of 0.5 g L−1 of both metal ions [126]. Another study showed the use of flexible polyvinyl conduit to create a filter containing Pseudomonas aeruginosa strain CMG156 biofilm for Cu(II) removal. It was discovered that the filter removed 85 percent of the Cu from the fluid. The biofilm of Escherichia coli maintained on kaolin was capable of removing several metals, and the order of biosorption was Fe(III) > Cd(II) > Ni(II) > Cr(VI) [80]. Similarly, Fe(III) adsorption was highest in the E. coli biofilm maintained on NaY zeolite, followed by Ni(II), Cd(II), and Cr (VI) [127]. The exopolysaccharides (EPS) are one of the most important components of biofilm. The functional groups in this biofilm polymer such as −OH, −COO, −NH, and C=O operate in sorption sites for a variety of heavy metals [128]. Due to the presence of these active and ionizable functional groups, heteropolysaccharides are frequently polyanionic when compared to homopolysaccharides. EPS present in biofilms that include molecules with surfactant or emulsifier qualities are used to bioremediate through biosorption [9]. Studies have found that a biofilm of Klebsiella sp. 3S1 entrapped on porous ceramic rings could remove 70 mg lead per gram of bacterial biofilm with 99 percent regeneration efficiency after four cycles of adsorption [129, 130]. The interaction of negatively charged functional groups on the EPS layer with cationic metals in this approach is independent of metabolism and energy. The exchange of ions between Pb(II) and the functional groups of EPS was used to detect the absorption of Pb(II) ions by Klebsiella sp. J1. Pb(II) also competes with K+ and Mg2+ for binding to the EPS matrix’s anionic groups [39]. The ionization state and adsorption characteristics of many functional groups of EPS are influenced by the pH of the adsorption medium. Because there are less H+ ions in the solution, metal ions adsorb more readily at higher pH. The role of EPS from Paenibacillus jamilae was detected in ferric ion precipitation. Furthermore, Fe(III) precipitation was shown to be greater at pH 6 than at pH 3 [131,132]. Toxic heavy metals form complexes with the anionic functional groups of EPS through electrostatic and covalent bonding. The carboxyl group gains a net negative charge in neutral solutions (pH 7) and interacts electrostatically with positively charged heavy metals to produce organometallic complexes.
Increased binding affinity for Hg(II) was also observed in the EPS of Bacillus thuringiensis PW-05. Similarly, EPS derived from Bacillus licheniformis has a greater affinity for Cu2+ and Zn than other bacteria [133]. Electrostatic contact is used to attach these metal ions to the functional groups of EPS components [134]. It is also found that the EPS from Paenibacillus jamilae has maximal complexation ability towards Pb (230 mg/g of EPS). The chemical and elemental makeup of a biosorbent may be determined through EDX analysis. The adsorption of Pb(II), Cr(VI), and Cd(II) by the EPS of P. pseudoalcaligenes NP103, Bacillus pumilus biomass, and Halomonas sp. cells was investigated using EDX spectrum analysis [128]. On the biofilm-EPS of P. aeruginosa JP-11, B. cereus BW-201B, and P. pseudoalcaligenes NP103, FTIR spectrum data showed the interaction of metal cations such as Cd(II), Hg(II), and Pb(II) with different negatively charged functional groups (−SH, P=O) [128]. FTIR analysis was used to show the trapping of several heavy metals on the biofilm-EPS of diverse bacterial species [134]. In experimental circumstances, the role of bacterial biomass and biofilm-EPS on the removal of hazardous metals has been thoroughly documented. However, more improvement and up-scaling of the methodologies are required to leverage their efficacy in the field investigation [134].
2.6. Genetically Engineered Microbes in Bioremediation
In the development of genetically engineered microbes (GEM) for bioremediation, four main techniques are being considered: (1) Chemical sensing, end point analysis, and toxicity reduction using bio affinity bioreporter sensors; (2) bioprocess creation, monitoring, and control; (3) affinity and enzyme specificity enhancement; and (4) route construction and regulation.
The selection of cellular factories is also a critical parameter to be considered. Among fungi, bacteria, and algal strains immense potentials has been reported for bacterial system attributed to several intrinsic features such as higher growth rate, containment, and ease in genetic manipulation. On the other hand, cyanobacteria and microalgae attributes to sustainability, and economic feasibility attributed to their photosynthetic activity and complex metabolic pathways resembles plant kingdom. The next criterion is to decide whether biosorption or bioaccumulation? The choice of biosorption and biosorption is primarily dependent on the heavy metal to be remediated, the concentration to be remediated, source for remediation, environmental conditions, other nutritional parameters, presence of other interfering molecules/ions, and so on. Irrespective of that, biosorption and bioaccumulation playing the primary role of heavy metal remediation, strategies are being framed to adsorb more number of heavy metals on the surface of cells and enhance the capability to accumulate the heavy metals by incorporating porters [135]. For example, the metal ion import systems (channels, primary active transporters, secondary carriers) are incorporated and overexpressed in certain bacteria to achieve higher uptake of respective heavy metals. On the other hand, further reduction/ remediation of heavy metals is being targeted heavily after bioaccumulation. For instance, enzymes and proteins that specifically reduces the heavy metal complexes are designed, incorporated in different organisms to enhance their remediation capabilities.
Incorporation of metal importers based on facilitated diffusion phenomena that depends on channel proteins were utilized for improving the uptake of arsenic and mercury ions [135]. In case of arsenic uptake, the Fps1 and glycerol facilitator from E. coli and Saccharomyces cerevisiae, respectively were used. Similarly for mercury uptake MerT/P transport system comprising MerF, MerE, MerC from Serratia marcescens was used [135]. These channels work based on energy independent simple diffusion process. In addition to that the periplasmic concentration of heavy metals are increased by over expressing the metal ion-selective porins in the cell wall, which govern the entry of all solute molecules in a cell. Under the category of secondary carriers, symporters are predominantly used for the uptake of cobalt, nickel and arsenic ions. For uptake of cobalt and nickel, NixA porter from Staphylococcus aureus and H. pylori were used as symporters. In case of primary active transporters, genetic engineering approaches are used to enhance the concentration of ABC transporters (type I and type II) in the cell membrane for effective uptake of heavy metals. Strains of Pseudomonas aeruginosa accumulate pseudopaline, which is a metallophore that assists in the uptake of heavy metals from the environment [136]. Similarly, S. aureus accumulates staphylopine, and Y. pestis accumulates yersinopine are other metallophores, which supports metal uptake. These intrinsic characteristic features of the wild type strains open up scope for genetic engineering in other strains for enhancement of specific metal uptake even at low concentrations [137]. Genes were produced to collect Cd2+ and release phytochelatins in the presence of a bacteroid-specific promoter (the nifH gene). Heavy metal degradation using genetically engineered microorganisms has piqued interest, as demonstrated by the use of Alcaligenes eutrophus AE104 (pEBZ141) for chromium degradation from industrial wastewater and the construction of a recombinant photosynthetic bacterium with simultaneous expression of the Hg transport system and metallothionein for Hg2+ degradation from heavy metal and wastewater Rhodopseudomonas palustris [3]. Cell surface engineering has also been gaining significant interest among researchers in which the metal specific peptides are expressed in the extracellular phase for enhanced adsorption and remediation. Numerous such cell surface peptide display systems are expressed in different microbial organisms and are exhaustively listed [138]. E. coli is one of the systems, where several cell surface engineering approaches are tested and validated. For instance, the metal binding protein EC20, siderophore binding protein, CueR for the binding of lead, iron, copper were tested, respectively [138]. A CadR gene from the wild type Pseudomonas putida strain was engineered in S. cerevisiae and showed ~6 folds increment in the binding efficiency for cadmium as compared to the wild type strain [78]. Even though several such studies are available significant understanding on the underlying phenomena and applications in real life scenario is still awaited.
2.7. Heavy Metals Removal from Soil
Co-contamination of soil with heavy metals and pesticides has resulted in major global environmental problems. The combination of heavy metals and insecticides further complicates the pollution issue. It is critical to clean up the soil that has been poisoned by heavy metals and pesticides. Bioremediation, which has been widely established in the remediation of combined pollution of heavy metals and pesticides, has the characteristics of high effectiveness (particularly for vast, low contaminated regions), cheap cost, easy availability, ecosystem safety, and high public acceptability [139]. Residual metals in soils have become a severe environmental concern as a result of their widespread use in activities. Organisms in polluted soils have several defensive strategies against pollutant toxicity. Bioremediation, which involves treating pollutants with microbes and/or plants, is a cost-effective and promising strategy for reclaiming co-contaminated soils [140].
Bioleaching technology is expected to witness some important advancements in the not-too-distant future, thanks to developments in microbiome and a better knowledge of biofilm ecology [118]. The use of acidophilic bacteria to improve the solubilization of solid phase heavy metals from sediment matrix is known as bioleaching or microbial leaching. The heavy metal components associated with iron or sulphur minerals in sediments are the primary targets of bioleaching. As a result, the most commonly used bioleaching microbes are members of the Proteobacteria (e.g., Acidithiobacillus, Acidiphilium, Acidiferrobacter, Ferrovum), Nitrospirae (e.g., Leptospirillum), Firmicutes (e.g., Alicyclobacillus, Sulfobacillus), Actinobacteria (e.g., they can oxidize iron or sulphide minerals, provide an acidic environment, and thereby solubilize immobilized heavy metals in water [141–143].
For example, in a uranium-contaminated sediment, the chemolithoautotrophic bacteria Leptospirillum was able to oxidize ferrous iron under an acidic situation with a pH range of 1.5–1.8, and Thiobacillus was able to derive energy from the oxidation of reduced sulfur and thiosulfate [144]. Furthermore, Aspergillus niger strain SY1 has been found to bioleach metals from polluted sediments and then reduce the bioavailability and toxicity of metals that remain in the sediments [145]. Abiotic variables such as the geochemical properties of metals in sediments, pH, and sediment particle size, as well as biotic factors such as microbial populations, metabolic activities and pathways, and microbial adaptability to minerals, all have a significant impact on bioleaching effectiveness. Depending on the geochemical parameters, indigenous heterotrophic Bacillus isolates might leach Cu, Zn, As, and Fe from polluted sediments, according to studies [145].
Total metal ion concentrations, chemical forms of the metals, and associated parameters such as redox potential are used to evaluate whether heavy metals are stimulatory or inhibitory to microbes. Temperature, pH, low molecular weight organic acids, and humic acids can all affect how heavy metals are transformed, transported, valanced, and bioavailable to microbes. With more protons available to saturate metal-binding sites, heavy metals tend to generate free ionic species at acidic pH values [9]. The adsorbent surface becomes increasingly positively charged as hydrogen ion concentrations rise, diminishing the adsorbent’s attraction to metal cations and hence increasing its toxicity. In the adsorption of heavy metals, temperature has a vital influence. The rate of adsorption diffusion over the exterior boundary layer increases as the temperature rises. Microorganisms’ activities are enhanced by a rise in temperature within a reasonable range, which boosts microbial metabolism and enzyme activity, speeding up bioremediation. The sorption sites, microbial cell wall structure, and ionization of chemical moieties on the cell wall all influence the stability of the microbe-metal combination. The substrate and a variety of environmental conditions influence the result of the degradation process [9].
Many of the investigations are conducted in laboratories under artificially controlled circumstances. The experimental parameters should be designed to mimic natural circumstances, such as climate, weather, and pollution levels (aging). More research is needed to validate bioremediation efficacy in real-world settings rather than pot trials. Field applications should be carried out in a variety of places. Potential novel hyperaccumulator species must be screened naturally or artificially with practical applications. Ornamental plants, landscape plants, bioenergy plants, and fast-growing plants are all possibilities for commercial usage. When organic pollutants are objectives for remediation, remediation solutions should be devised for both the organic pollutants parent and their metabolites, not simply the organic pollutants parent [148]. We should be aware of the fact that some organic pollutants’ metabolites have a lengthy half-life or are even more harmful than the parent pollutants. To minimize secondary contamination caused by the death of organisms or changes in the environment, the plants and microorganisms employed for bioremediation of polluted soil must be treated thereafter. Bioenergy generation, phytomining, papermaking, wood processing, and other aspects of plant biomass post-harvest management might be explored [147]. Non-usable biomass should be disposed of as hazardous trash. More study is needed into the mechanisms of various contaminants interacting with bioremediation and/or phytoremediation processes. Meanwhile, further study is needed to understand the processes of plant-microbe, plant-plant, and microbe-microbe interactions under pollution stress. Soil contamination assessment techniques prior to remediation, as well as efficacy evaluation methods after remediation, should be more precise, integrated, intelligent, energy efficient, and quick [148].
3. Future Prospects
The unrestricted dumping of industrial effluents into agricultural fields or bodies of water enhances the likelihood of their entering the food chain via crops and aquatic animals, and then bioaccumulating. Bioremediation strategies suited for various environmental circumstances, both in situ and ex situ, have been explored and suggested. The use of biological agents in the design, development, and implementation of these approaches necessitates careful selection. For bioremediation, extensive research is being conducted utilizing specialized strains of microorganisms. Microorganisms carry out redox reactions, and as a result, metal mobilization/immobilization has an influence on bioremediation processes. By generating oxidized Mn, the Mn(II)-oxidizing Bacillus sp. strain indirectly oxidizes and mobilizes Cr(III) into bioavailable form of Cr(VI). Heavy metal bioremediation is more efficient when many microbial strains are used simultaneously rather than just one. Advances in genetic engineering and optimization approaches indicate that these technologies have a bright future. For some pollutants, genetically engineered microbes may have a superior bioremediation capability. Agricultural and industrial waste biomass, such as sugarcane bagasse, coconut shell waste, rice husk, and beer waste yeast, are also being explored as bio remediators on a lab/commercial scale. After different physical and chemical alterations, the biosorption ability of diverse biosorbents are increased, and further study is needed before these biosorbents may be used commercially across sectors. The bioremediation strategy necessitates a comprehensive and all-encompassing process for developing systematic, practical, and long-term methods that can be easily modified for any situation. Furthermore, collaboration at all levels, including research groups, the general public, governmental institutions, and industry, is critical.
4. Conclusions
Heavy metal contamination can come from both man-made and natural sources. Heavy metals should be removed from the environment due to their non-biodegradable and harmful nature. Industrial wastewater spilled into the environment, such as soil and rivers, need urgent government involvement, ongoing monitoring, and rehabilitation using proper technologies. Traditional treatment methods have limits and should be replaced with more efficient, cost-effective, and environmentally acceptable alternatives like bioremediation using biological agents. A key challenge is selecting an adequate bio-sorbent in terms of efficiency and cost. Microorganisms carry out redox reactions, and as a result, metal mobilization/immobilization has an influence on bioremediation processes. Microbial activities such as growth, colonization, and microbial biofilm development for remediation are influenced by metal-microbe interactions.
Notes
Conflict of Interest
The authors declare that they do not have any conflicts of interest.
Author’s Contribution
P.J. (PhD student) contributed equally to the manuscript conceptualization, preparation and wrote the manuscript. C.D. (PhD student) contributed equally to the manuscript conceptualization, preparation and wrote the manuscript. R.V. (Professor) edited, corrected the manuscript. M.M. (Assistant Professor) revised, edited, corrected the manuscript.