Prevalence of microplastics, antibiotic resistant genes and microplastic associated biofilms in estuary - A review
Article information
Abstract
The estuarine ecosystem is under threat due to the addition of different anthropogenic pollutants. Among the various pollutants, microplastics(MPs), and antibiotics play a significant role in affecting estuarine organisms and human health by transport through the food chain. In the estuaries, microorganisms, including pathogens, colonise microplastics by the development of biofilms. Estuaries have long been home to MPs, served as novel hubs for the transmission of Antibiotic Resistance Genes (ARGs). As a result of their continual interactions with a variety of aquatic creatures, MP-associated bacterial communities will eventually present a transfer opportunity to organisms that consume MPs. To comprehend the presence of microplastics and ARGs in the estuaries, a thorough review is necessary. This review discusses the various sources of MPs and antibiotic pollution, as well as the transport of ARGs via biofilm formed on MPs. Furthermore, the factors affecting biofilm formation in estuaries are reviewed. In addition, the transport of ARGs by microbial populations within biofilm is discussed. The ARGs are transported into the food chain, which will be a threat to human health. Hence, the occurrence of antibiotic-resistant genes in fish is discussed.
Abstract
Graphical Abstract
1. Introduction
Estuaries are highly productive and dynamic areas at the transition between rivers and the marine environment. Tributaries transport nutrients into estuaries and transform them into highly productive ecosystems [1,2]. Further, estuaries are vital resources for providing food and habitat for fish and shellfish, food for human consumption, areas for tourism and recreation, and a host of ecosystem services [3–5]. Currently, the estuarine ecosystem is highly polluted [2] by several point and nonpoint sources, including agricultural runoff, wildlife excrement, septic tanks, and sewage discharges [6,7]. Because estuaries are coastal transition zones between freshwater and seawater, pH, temperature, geochemical condition, and salinity vary on a relatively short spatial and temporal scale [8]. The geomorphological and physical-chemical characteristics of estuaries favour the deposition of organic matter and man-made contaminants [9]. The prevalence of low energy and currents from the waves in estuaries makes the pollutants accumulate in sediments instead of dispersing [10]. The types of pollutants found in estuaries are heavy metals [11, 12], radionuclides [13], and pathogens such as bacterial pathogens [14], protozoa [15], enteric viruses [16], and fungus [14]. Furthermore, antibiotics, antibiotic resistant genes, and MPs are regarded as important emergent pollutants in the estuarine ecosystem due to their widespread distribution, persistence, and impact on the environment and human health [17].
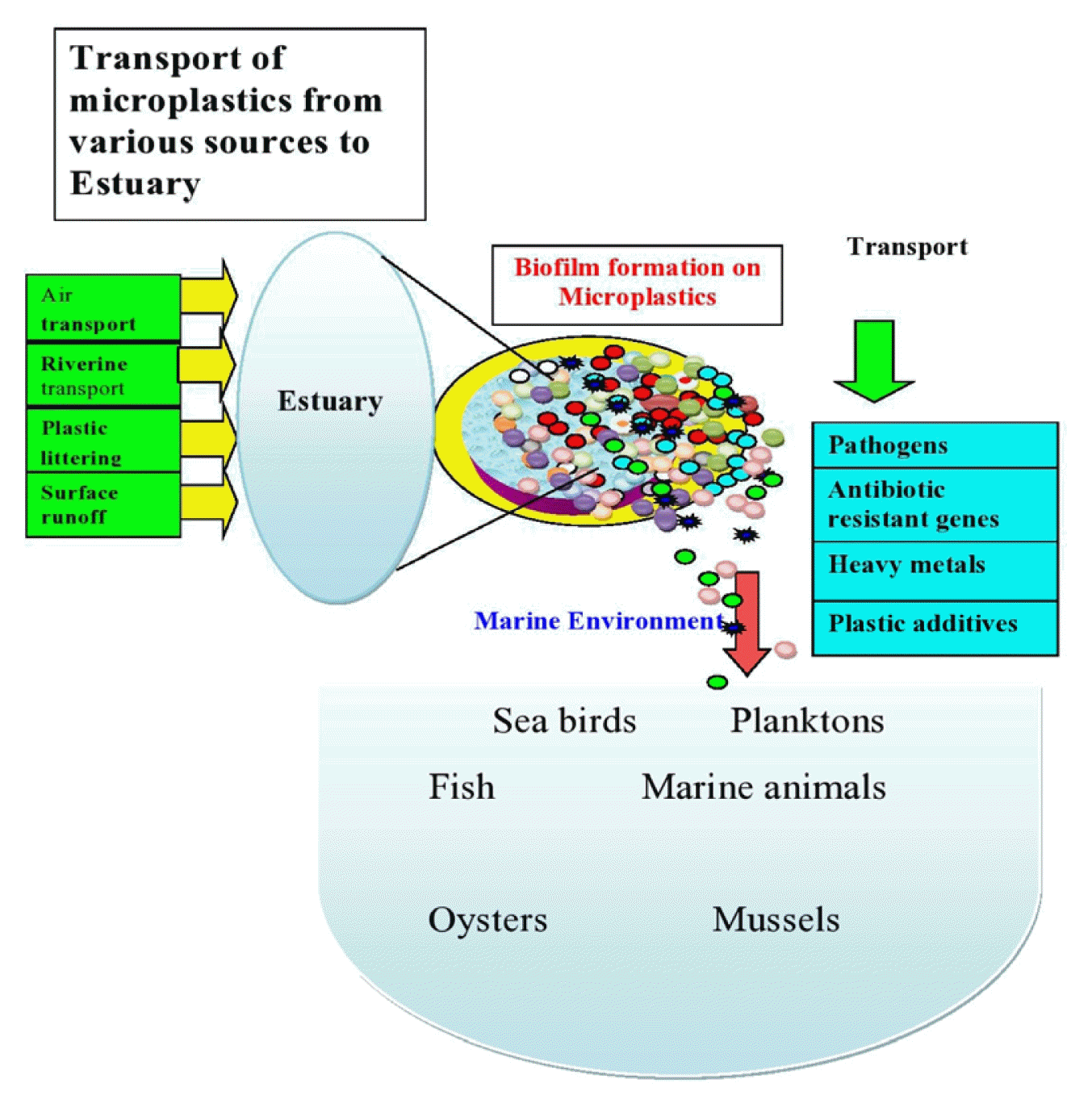
Estuaries also play an important role in the transport of pollutants from land to sea [15]. According to research, contaminants are moved from estuaries to the oceans through biofilm that develops on substrates such as MPs [18]. MPs can adsorb toxic pollutants such as antibiotics and heavy metals, both of which have been related to antibiotic resistance [19]. MPs could thus act as vectors for pathogens and antibiotic resistance genes in the aquatic environment [19]. MPs release absorbed toxic pollutants in the marine environment [20] and are consumed by marine biota, including seafood species [21]. Pollutants are indeed being transported into the food chain, which has emerged as a major issue in this decade. As a result, the primary goals of this review are to emphasise the following to highlight the current state, knowledge gaps, and future research directions: (1) the presence of MPs, antibiotics, and ARGs in estuaries; (2) the formation of biofilm on MPs in estuaries; (3) factors influencing biofilm formation on MPs; and (4) the transport of ARGs within the biofilm microbial community.
2. Antibiotic Pollution in Estuaries
Antibiotics are used to treat bacterial infections, namely urinary tract infections, sepsis, sexually transmitted diseases, and diarrhea [22]. It is observed that a high rate of resistance has been developed to the antibiotics used to treat these diseases [23]. Antibiotic resistance is accelerated by the overuse or misuse of antibiotics by people [24]. Antibiotic and ARG prevalence is being investigated in agricultural soils [25], rivers [26], and oceans [27]. Besides that, antibiotics pollute the estuaries [28]. Wastewater treatment plants (WWTPs) [29], leachate treatment plants [30], animal husbandry facilities [31], organic dairy farms [32] aquaculture industry [33], poultry farms [34], sewage contamination [35], household and hospital wastewater [36], and biology laboratories [37] are major sources of antibiotics in estuaries. Because antibiotics are not completely metabolised by the human or animal body, functional compounds can enter the aquatic ecosystem via the waste products of humans or animals that have consumed the antibiotics [38]. Furthermore, the types and concentrations of antibiotics in aquatic systems vary globally depending on antibiotic consumption and use [39]. Furthermore, the chemical stability and partition coefficient of antibiotics influence their presence in the aquatic environment [40]. The concentration of antibiotics is very low, and the design of water treatment plants is insufficient to remove the antibiotics [41]. As a result, even treated effluents from WWTPs can be a source of antibiotics in the aquatic environment [42].
Antibiotic resistance is a global health concern; infections with resistant pathogens kill at least 700,000 people each year [43]. According to WHO (2018), pathogenic bacteria resistant to common antibiotics are found in human and veterinary systems. There is an increase in global annual deaths due to antibiotic resistance, which could be increased to 10 million by 2050 [44]. Antibiotic pollution has been found to be more severe in the spring, summer, and winter seasons than in the autumn [45]. In general, antibiotic concentrations in sediment samples were higher than in water samples [46]. Many laboratory scale studies have proven that the presence of antibiotics at concentrations as low as nanograms per millilitre has been shown to promote the development of resistance [47]. Due to the harmful effects of low levels of antibiotics in the environment, antibiotics are now considered an important environmental pollutant [48]. The study conducted in the Yantze river estuary; China showed that the concentrations of antibiotics varied with location [49]. In addition, the highest antibiotic concentration was observed around river discharge and sewage outfall during the January month. Furthermore, there was a positive correlation between total antibiotic concentration and dissolved organic carbon (DOC) concentration [49]. The prevalence of the highest concentration in sediment samples might be due to low flow conditions and low temperatures [50]. The antibiotics, namely tetracycline, sulfaquinoxaline, enrofloxacin, and thiamphenicol, were examined in the Yangtze Estuary and nearby coastal areas [50]. There was a positive correlation between quinolone concentration and Total Organic Carbon (TOC), indicating the significant role played by TOC for the increased accumulation of antibiotics in sediments [50]. In the Jiulong River Estuary, antibiotics such as sulfonamide, quinolon, and chloramphenicol were examined [51]. In India, a study conducted in the Cochin Estuary revealed the occurrence of Vibrio parahaemolyticus which was found to be resistant to antibiotics, namely enrofloxacin, nitrofurantoin, trimethoprim, sulphamethoxazole, streptomycin, amikacin, and nalidixic acid indicating the presence of aforementioned drugs [52]. Further, it was evident that the antibiotic resistant genes were present in plasmids [52]. The abundance of salfonamide-resistant E. coli was reported in Liaohe and Daliaohe Estuaries in China, suggesting the population was higher during the dry season when compared to the wet season [53]. Furthermore, there was significant attenuation of antibiotic-resistant E. coli during movement between land and sea, explaining the role of the estuary as a natural attenuator in reducing the ARGs during the transport of antibiotic-resistant bacteria from rivers to the ocean [53]. In general, ciprofloxacin increased ARG types in biofilm and induced the growth of dominant bacteria belonging to phyla such as proteobacteria, actinobacteria, and planctomycetes [54].
3. Microplastic Pollution in Estuaries
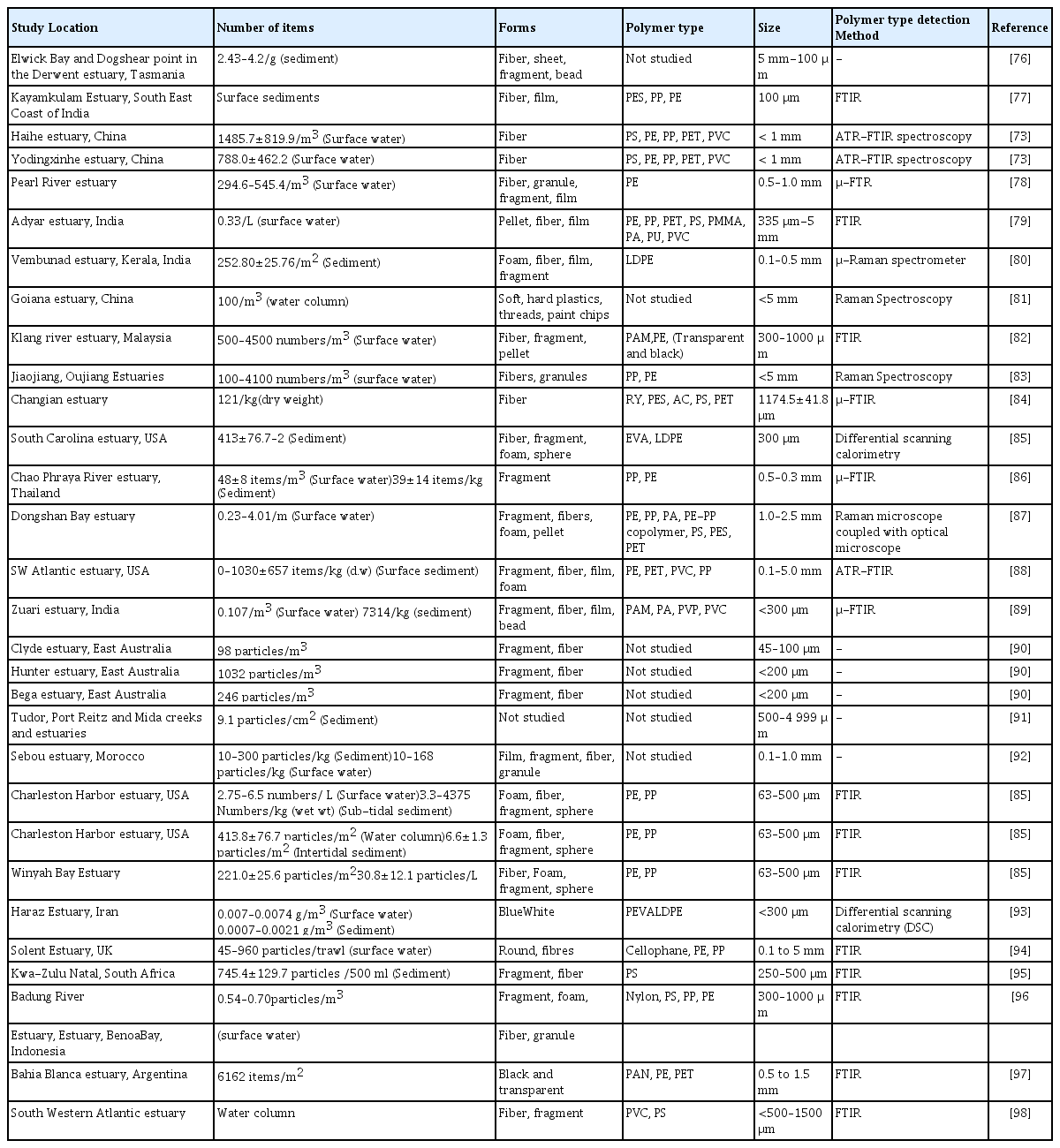
Microplastic pollution is a serious threat to the environment and is considered an emerging contaminant of concern. MPs are heterogeneously mixed plastics that are less than 5 mm in diameter. MPs are distributed in different forms, namely plastic fibres, granules, and pellets [55]. The distribution of MPs is observed from the poles to the equator [56]. Plastics enter the environment primarily as a result of human activity [57, 58]. The important sources of plastic pollution are agricultural mulching [59], sewage sludge disposal [60], wastewater treatment plants [61], informal landfills [62], and biowaste compost [63]. MPs are found in a variety of terrestrial environments, including home and garden soils [64], flood plain soils [65], remote high mountain soils [66], and industrial soils [67], as well as surface and deep ocean waters [68], lakes [69,70], mangroves [71], shorelines [72], and estuaries [73–75]. The studies conducted to examine the distribution of different sizes, forms, and types of MPs in estuaries across the world are presented in Table 1.
According to the investigations, estuaries were filled with MPs in the form of fibre, fragment, foam, film, and pellets [89–93]. Unlike in surface water, MPs from sediment samples were predominantly in the 0.05–0.3 mm size range, making them easily ingestible by aquatic organisms [86]. The shallow sediment contained more microfibers, and the rate of microfiber deposition in the sediment was strongly related to the expected change in production in the study area based on annual plastic production [76]. Furthermore, seawater inundation in the estuary and the distance of sampling points from the coast had a significant influence on the distribution of fibres and films in estuary sediments [77]. The most common polymer types found there were PE, PP, PVC, PA, PAM, and PES [96–98]. However, [78] suggested that MPs carried by upstream river water to the estuary were an important source of MP pollution, and the abundance of MPs varies greatly across regions due to differences in human activities as well as hydrological and hydrodynamic conditions. However, the studies that have been done to look at how different kinds and forms of MPs are distributed in estuaries are insufficient[99]. Because of the vegetation and subsequent flocculation, estuaries can filter pollutants more effectively [100]. Many estuary components, such as coastal wetlands like mangroves, salt marshes, lagoons, and tidal and subtidal flats, act as sediment and pollutant sinks [101]. Because of the extensive weathering nature of microplastics in the environment, their virgin nature is altered, and thus their physiochemical characteristics are altered [102]. Furthermore, changes in crystallinity, specific surface area, and oxygen functional groups cause the sorption behaviour of MPs to change [103]. Furthermore, degradation can be confirmed by a loss of gloss and/or an increase in haze caused by oxidation and cleavage of polymer chains[104]. Semicrystalline polymers, such as PE, experience an increase in crystallinity (and brittleness) as a result of chain scissions in amorphous regions [105]. Photooxidation, on the other hand, causes cracks and increases fragmentation [106].
4. Biofilm Development in Estuaries
In the aquatic environment, bacterial phenotypic plasticity is responsible for bacterial adaptation to different environmental conditions [107,108] which is achieved through the formation of multi-species biofilms, which enables interspecies interactions responsible for ecological processes [109]. Biofilm is commonly composed of microbial cells and their metabolic products, such as extracellular polymers and inorganic precipitates [110]. Bacterial biofilm is thought to be an emerging form of bacterial life in which communal life differs significantly from bacteria that live as free-living cells [111].
Biofilm protects microbes and allows them to survive and thrive in harsh environments. Furthermore, microbes in the biofilm are less susceptible to antibiotics and disinfectants when exposed to them [112]. Bacteria grown in biofilms, on the other hand, can form intricate and complex structures, such as channels that allow nutrients to circulate. The structure of biofilm influences gene expression [113]. Biofilm formation occurs on a variety of substrates, including living tissues, indwelling medical devices, piping in industrial or potable water systems, and natural aquatic systems. Marques et al. [114] investigated the formation of biofilms on hydrophilic surfaces such as stainless steel and glass. The bacterial community in estuaries is dependent on precipitation, according to a study conducted in the Bilbao estuary [2]. Freshwater bacteria from the Comamonadaceae and Spingobacterioaceae were abundant during high precipitation periods. During low precipitation periods, however, marine bacteria from the Rhodobacterales and Oceanospirillales predominated [2].
4.1. MPs as Carriers of ARGs
Estuaries, serving as links between land and ocean, harbour numerous microbes that are relatively highly active because of the massive terrigenous input of nutrients [115]. Furthermore, ARGs such as tetracycline, sulfonamides, AmpC β-lactamase, and the class-1 integron gene were found in the leachate treatment plant effluent [116]. ARGs such as sul1, int1, and sul2 were prevalent in the receiving aquatic system. Furthermore, research has found a relationship between ARG concentration and heavy metal concentration [117]. It was observed that the genes encoding antibiotic resistance and heavy metals were found to be located in the same position on chromosomes or mobile genetic elements, which could facilitate the acquisition of multiple resistance genes by bacteria [118]. There was a strong positive correlation between vanadium and gene tetM [119]. Vanadium has physiologically negative effects on a variety of microbes [120], and it can cause bacterial resistance to vanadium via efflux pump tricarboxylic acid cycle enzymes and accelerate horizontal gene transfer [120]. As a result, it is intriguing to speculate that this resistance could confer resistance to ARGs in bacterial communities in the aquatic environment [119]. Metal absorption would also be aided by interactions between metal ions and charged or polar regions of the MP surface, or by non-specific interactions between neutral metal-ionic complexes and the hydrophobic surface of MPs [121].
Since plastic has a longer half-life when compared to natural materials, the plastispheres or microporous structures formed on microplastics provide a favourable condition for attachment and subsequent formation of biofilm by microorganisms [122]. It is evident from the studies conducted so far that weathered polystyrene foams are acting as carriers of antibiotics in the environment [117].
Salmonella sp. and E. coli were the bacterial species that had the ARGs AmpCo, blan-TEM-1, bla-CTX-M, VIM-1, NDM-1, florR, tetG, and mcr-1 [123]. Additionally, the penetration into the biofilms is determined by the type of antibiotic [124]. In contrast to ciprofloxacin, which readily entered the biofilm created by wild-type Klebsiella pneuminiae, ampicillin did not [124]. The major enzymes involved in nitrogen cycling (the anammox, denitrification, and nitrification processes) are altered by antibiotics in aquatic systems, which has an impact on ecosystem performance [125]. Other strains could quickly acquire the plasmid-encoded virulent genes and antibiotic-resistant genes [126]. Table 2 provides a summary of the studies done to determine the occurrence of ARGs in estuaries. Additionally, there have been relatively few studies that look into the sources, characteristics, and distribution of ARGs globally (Table 3). The most common antibiotic resistance genes found in estuaries were Sul, Tet, and Qnr [127, 128]. The antibiotic resistance found in the bacterial isolates, combined with the longevity of plastics in the aquatic environment, suggests that biofilm on plastics has the potential to harbour pathogens and facilitate horizontal gene transfer [129]. The studies show that more research is needed to investigate the distribution of different types of ARGs in estuaries. This could allow us to take appropriate steps to prevent the spread of ARGs into estuaries.
4.2. Biofilm Development on MPs in Estuaries
There are more than 1200 estuaries in more than 120 countries and territories worldwide, including lagoons and fjords [152]. However, the research on the development of biofilms on microplastics was only done in a few numbers of estuaries around the world. Plastic biofilms are an important site for potential pathogen colonisation and horizontal gene transfer since they are long-lasting [129]. The studies carried out to look at the presence of biofilm on MPs in various estuaries around the world are shown in Fig. 1.
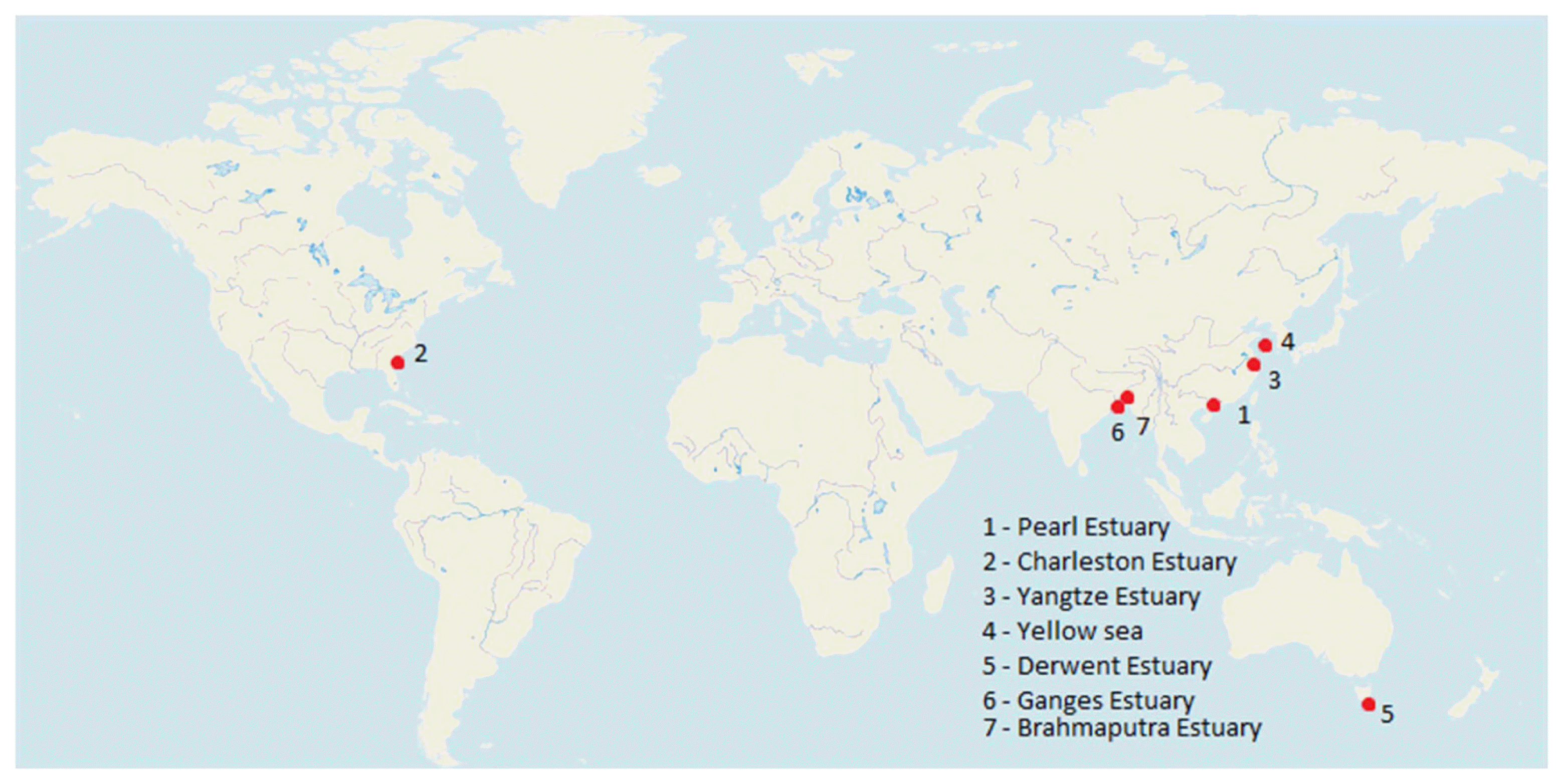
Studies conducted to examine microplastic based biofilm formation in estuaries across the world (The red dots indicate the estuaries).
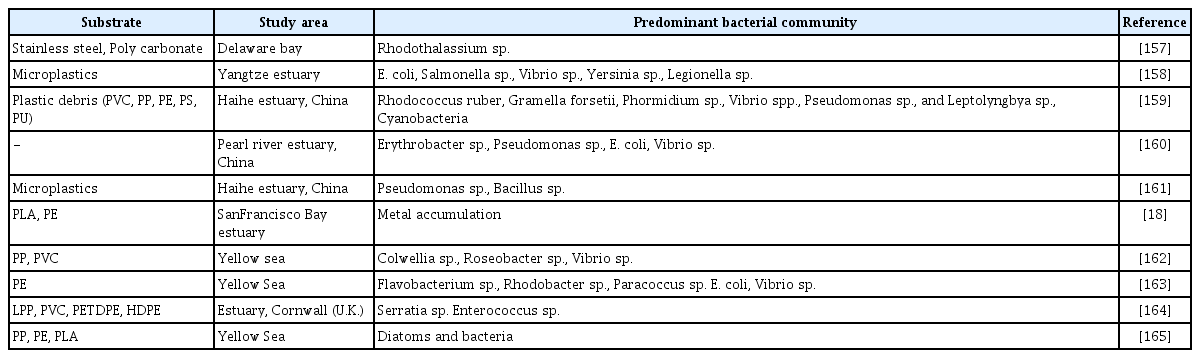
The environmental conditions, the bacterial genome, and the components embedded in the biofilm matrix all influence the structure and components of biofilm [153]. The optimal temperature for Salmonella sp. biofilm formation was found to be 25 to 42°C and pH values of 7–8 [154]. A carbon source is also required for biofilm formation [154] Furthermore, the mechanisms involved in biofilm formation differ depending on the bacterial strain and environmental conditions [155]. Surface chemistry and surface energy are vital properties for microorganism attachment. Furthermore, four distinct three-dimensional architectures were observed when immersing stainless steel and polycarbonate surfaces in a biofilm in an estuary [156]. For example, mushroom like structures, homogeneous, hairy biofilms with horizontal fibres and heterogeneous tousled biofilms were formed by Flavobacterium sp. 112003, Roseobacter sp. IV3009, Shewanella sp. IV3014 and Roseovarius sp. VA04, respectively [156]. The antibiotic resistance detected among the isolates coupled with longevity of plastics in the aquatic environment suggests biofilm on plastics have potential to pathogens and horizontal gene transfer [129]. The bacterial community structure of biofilm developed on different types of MPs in estuaries is shown in Table 4.
It is evident from previous studies that MPs can readily release hazardous pollutants like DDT, polybrominated diphenyl ethers, and other additives into the aquatic environment, thereby elevating the concentration [166]. Also, MPs are transporting organic pollutants into the environment [167]. In the biofilm, a variety of microorganisms with varying redox potential requirements occur in close proximity [168]. In general, preferential attachment of cells during biofilm formation is achieved by the different topography of the substrates, namely, irregular surfaces, etching surfaces, and grooved surfaces [169], when compared to flat surfaces. Furthermore, it is clear that biofilm formation is strongly influenced by environmental nutrient concentrations, such as nitrogen [170] and carbon sources (mannose and trehalose) [171]. In general, sessile microorganisms in biofilms have high environmental stability, more opportunities for interactions such as horizontal gene transfer and co-metabolism are resistant to physical disturbances and antimicrobial compounds [172]. During biofilm formation, the initiation of corrosion reactions by microorganisms is achieved by the formation of concentrated cells, the production of corrosive metabolites, the dissolution of protective layers, the uptake of electrons directly from the substrate, and the production of non-protective layers [173]. In addition, the high population densities, and proximity of cells in biofilms increase the chances for genetic exchange among the bacterial species, converting biofilms into hot spots of antibiotic resistance [174].
4.3. Factors affecting MP associated Biofilm Formation
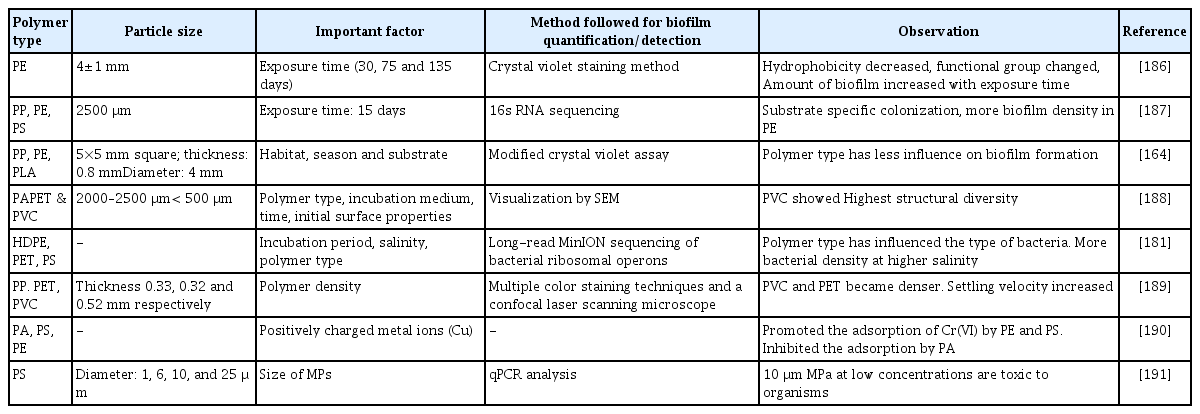
The important factors affecting MP associated biofilms are size and shape of MPs, hydrophobicity, plastic additives, presence of phenolic compounds and heavy metals [162, 175, 176]. The different factors affecting the biofilm formation on MPs and the effect of biofilm on MPs is illustrated in Fig. 2.
Few studies demonstrated that the microbial community structure of biofilm formed on MPs is influenced by polymer type. Debroas et al. [177] examined that PS, PE, and PET plastic particles were dominated by Alphaproteobacteria and Gammaproteobacteria while Burkholderiales dominated on PE MP biofilms. Further, the early colonizers on PE, and PS MPs were found to be Vibrio sp. [178]. However, the dominant PE colonizing microbial community varies during biofilm formation [129]. The biofilm structure and function depend on microbial growth rate and the type and level of energy sources [179]. In general, microorganisms colonized in the upper regions of the biofilm are more active due to greater accessibility to nutrients and capable of discharging metabolic waste products compared to cells at the bottom region [180]. The biofilm developed on three types of MPs prevalent in the environment namely HDPE, PET, and PS was examined. The virgin MPs were incubated for a period of 31 days with water collected along freshwater - estuarine gradient of the Raritan River in New Jersey, USA. The study revealed that salinity and polymer type had impact on formation of biofilm in which colonization of distinct microbial community was achieved. One of the most abundant MP colonizing bacterial species was found to be Limnobacter thiooxidans [181].
The quantum of chemical energy possessed by plastics is greater, but it is unclear how biofilm formation and microbes facilitate the degradation of plastic debris [182]. There is a relationship between biofilm formation and the physicochemical characteristics of polymers. The characteristics of the plastics, namely compression, crystallinity, surface chemistry, hydrophobicity, and surface topography of plastic particles, determine the microbial community structure in the biofilm [183]. Spingobium, Novospingobium, and uncultured Planctomycetae were the predominant microbial types colonised on polyethylene MPs [183]. Further, significant changes in crystallinity of PE, stiffness of PP, and compression of PS were observed due to biofilm formation [183]. The PE degrading strain of the actinomycete Rhodococcus ruber showed laccase activity and reduced the molecular weight of PE [184]. However, the biofilm formed in the LDPE-Titania environment increased hydrophobicity by the different bacterial species, namely Pseudomonas aeruginosa (CA9), Burkholderia seminalis (CB12), and Stenotrophomonas pavanii (CC18) [185]. The factors influencing the colonisation of microorganisms in biofilms developed on MPs are summarised in Table 5.
Plastics are degraded by the microbial community colonised by plastic particles [185]. Microorganisms achieved preferential degradation of amorphous regions and non-preferential degradation of plastic surfaces in general [192]. Vaksmaa et al. [183] discovered the presence of various bacterial communities on plastics belonging to the orders Flavobacteriales, Alteromonadales, Chitinophagales, and Oceanospirillales. Furthermore, Ideonella sakaiensis [193] degraded PET, Alcanivorax borkumensis [194] degraded low density polyethylene (LDPE), Zalerion maritimum [195] degraded PE, and Aspergillus sp. [196] degraded PE (HDPE). On the surface of the PE MPs, a wide range of biofilm morphological types were observed, including coccus, rod-shaped, and disc-shaped bacterial cells, interwined filaments, and a dense layer of EPS [186]. Similarly, the hydrophobicity of the PE decreased with exposure time. It could be because the surfaces of the MPs absorb organic and inorganic nutrients from water in a matter of hours [186]. This results in the formation of a conditioning film, which can quickly attract microorganisms and facilitate the utilisation of nutrients on said surfaces [186]. The larger the contact angle, the greater the hydrophobicity of the solid surface. It was found that the contact angles for PA, PET, and PVC were 70°, 81°, and 87° respectively. Hydrophyllic surfaces are typically associated with contact angles less than 90° [188]. The PVC with the lowest negative ζ-potential showed the highest structural diversity after incubation in seawater, indicating that the structural diversity is additionally dependent on the incubation medium [188]. Temperature had an impact on the formation of a biofilm on MPs, which was connected to metabolic activity and enzyme activity [175].
4.4. MPs as Carrier of Antibiotics and ARGs
When compared to virgin materials, biofilm formed on MPs can absorb more pollutants. Furthermore, MPs in biofilm can transport pollutants and microorganisms to aquatic ecosystems [167,197]. Besides that, the toxic effect of plastics can be determined by particle size and surface modification. Nanoplastic-induced oxidative stress can reduce biofilm enzyme activity [176]. Studies confirmed that the surface properties of LDPE microplastics influenced the biofilm communities that formed on them. However, pathogens were more prevalent in biofilm colonised on LDPE microplastics [198].
The occurrence of antibiotic resistance genes was studied by researchers in estuaries. Biofilms are considered as hot spots of antibiotic resistance [199] due to the exchange of genetic elements among bacterial species, which is achieved by high population density and proximity of cells [174]. When susceptible microorganisms in biofilms are exposed to antibiotics in the environment, they become resistant to antibiotics due to selective pressure, chromosomal mutations, and horizontal gene transfer. It is understood from previous studies that ARG transfer is mainly achieved by horizontal gene transfer [200]. The genes responsible for antibiotic resistance are located on mobile genetic elements present in bacteria, namely plasmids, transposons, and membrane vesicles, integrons and membrane vesicles [201] cause horizontal gene transfer [202]. Some bacteria carry plasmids, which provide multiple antibiotic resistance and allow them to survive in harsh environments [203]. Antibiotic-resistant bacteria that have detached from biofilms can spread in the environment and affect human health [199]. Furthermore, antibiotics and ARGs influence nutrient cycling in aquatic systems by altering bacterial community related to biogeochemical cycling Zeng et al. [204]. The sources of ARGs and horizontal gene transfer between microorganisms colonized within biofilm developed on MPs are shown in Fig. 3.
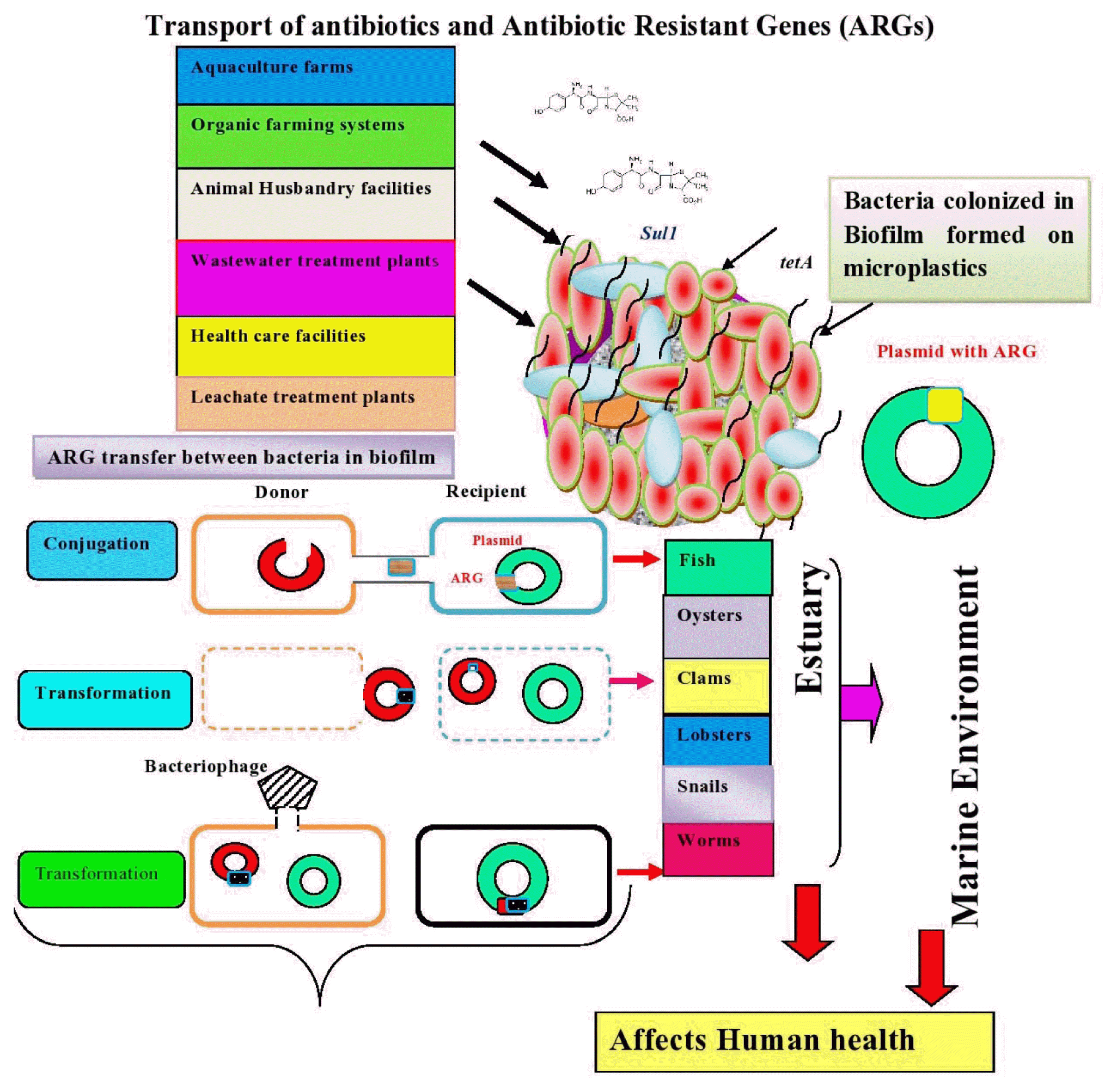
Schematic showing sources of antibiotic resistant genes and horizontal gene transfer within biofilm developed on microplstics in estuaries.
The mobility of ARG transfer between cell donor and recipient is achieved by conjugation, transformation, and transduction. However, intracellular transport is facilitated by integrons and transposons [205]. The study conducted by Ma and Bryers [206] revealed that the bacteria in biofilm sense the antibiotics they are resistant, which enhances the spread of antibiotic resistant. It is suggested that high surface/volume ratio of biofilm favors the efficient transfer of genes between or within the microbial populations [207]. Many studies were conducted to examine the conjugation mediated horizontal transfer of ARGs by bacterial species namely E. coli [145] and Pseudomonas putida [208]. The transformation was another way transfer of genetic element from donor to recipient in Bacillus subtilis and Acinetobacter baylyi ADP1 [209]. In addition, gene transfer is achieved by transduction in which E. coli and Flavobacterium sp. were the donor and recipient respectively [210]. The study conducted by Solheim et al. [211] revealed that phage mediated stx-gene transfer was facilitated between E. coli O103:H25:1199 (recipient) and E. coli C600 731 within biofilm.
MPs are the niche for many microorganisms and provide microhabitats, especially for opportunistic pathogens. In the aquatic systems, MPs selectively enrich ARGs within the biofilm [212]. Interactions between MPs and ARGs are mediated by van der Waals forces, electrostatic forces, and π- π - interactions between MPs and ARGs [212]. It was observed that the bacterial community on MPs possessed higher average path length, clustering coefficient, and modularity compared to those in water samples [167]. The MPs examined in the rivers Jiang and Zhejiang in China possessed a relative abundance of integrin-integrase gene classes 1 and 2, which facilitate horizontal gene transfer [167]. But limited studies have proven that the genetic materials collectively known as resistomes provide molecular function that protects the bacteria against many antibiotics [213]. He et al. [214] demonstrated that tetracycline resistant profiles and ARGs were observed in Listeria spp. isolated from catfish fillets. However, Aeromonas sp. isolated from the gut, liver, skin, mucus, gills, and flesh of rainbow trout harboured aadA, sul1, and tet genes [215]. The ARGs, namely blaDHA, blaACC, tetV, tetM, mdtE, mexF, vanTG, vanWG, vgaB, msrA, and mphA, were examined in fillets of rainbow trout fish, and the antibiotic resistance genes were carried by Carnobacterium sp. and Psedomonas sp. [216]. Miller, Harbottle [217] observed severe mortalities in larvae caused by Vibrio harveyi resistant to trimethoprim-sulfamethoxazole, chloramphenicol, erythromycin, and streptomycin. In addition, Aeromonas sp. colonised in carp harvested from pond water was found to be resistant to β-lactam and lincosamide antibiotics [218].
The antibiotic resistance genes (ARGs), namely tetA, tetO, tetQ, tetW, sulI, sulII, and blaTEM-1, and class 1 integrase (intI1) in the range of 9.4 ×10−6 − 1.6×10−1 and 6.7×10−5 − 5.2×10−1 gene copies per 16S rRNA gene, respectively, were observed in the guts of four major Chinese freshwater carp (i.e., silver carp, grass carp, bighead carp, and crucian carp) from food retail markets. The antibiotic resistance genes were present in Acidobacteria, Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes, and Proteobacteria [219]. In addition, V. harveyi isolated from a shrimp farming facility was found to be resistant to ciprofloxacin, penicillin, rifampicin, and vancomycin [220].
5. Conclusions and Perspectives
Based on the preceding discussion, it is possible to conclude that estuaries all over the world are under threat from antibiotics, antibiotic resistant genes (ARGS), and MP pollution. Agricultural soils, animal husbandry facilities, sewage treatment plants, and aquaculture systems are major sources of ARGs and microplastics. MPs found in estuaries are an important substrate for the colonisation of various microorganisms with ARGs. Furthermore, the polymer type influences the microbial community structure in biofilms formed on MPs. Horizontal gene transfer also allows ARGs to be transferred between microorganisms within a biofilm. Furthermore, the prevalence of antibiotic resistance in fish is investigated, resulting in resistance to pathogen-induced infections. Effective wastewater treatment facilities are suggested to prevent the entry of antibiotic, ARG, and MP residues into estuaries. Furthermore, prescribing antimicrobials by human and animal health care professionals based on guidelines will reduce antibiotic overuse, thereby controlling antibiotic accumulation in the environment.
The following study areas could be investigated in light of what is currently known about MP-related biofilm formation in estuaries: (i) The size, colour, and polymer type of MPs are all variable in aquatic environments. MP detection using a single detection technique is challenging. To more accurately detect MPs, multiple detection approaches are needed (ii) There has been little research done to date on the detection of ARGs in aquatic environments like estuaries. If it doesn’t get enough attention, ARGs will move up the food chain and have negative effects. It is crucial to recognise the different ARGs that take place in estuaries. (iii) There isn’t much information currently available to pinpoint the origin of ARGs in estuaries. As a result, source identification is paid greater emphasis, and the appropriate action is required. (iv) It is crucial to think about the bacteria that MPs have colonised. This is accomplished by utilising cutting-edge technologies, like as genomes, metagenomics, proteomics, and metabolomics, to understand more about the formation of biofilms and the biodegradation of MPs. (v) In estuaries, the mechanism of biofilm development on MPs of various sizes and types is poorly understood. It is necessary to pay more attention to how physical, chemical, biological factors, and MP interactions influence biofilm formation and the movement of ARGs and hazardous contaminants through the trophic levels in estuaries. (vi) ARG occurrence in estuaries is still out of control. For the protection of aquatic environments, it is necessary to track their behaviour and develop a risk assessment approach.
Acknowledgments
The authors thank the Centre for Environmental Studies, Anna University, Chennai, Tamil Nadu, India, for providing research facilities. The authors would also like to thank the two anonymous reviewers who provided helpful suggestions for revising the manuscript.
Notes
Author contributions
A.M.S (Assistant Professor (Sr. Gr.)) conceived the idea and wrote the manuscript. G.D. (Assistant Professor Sl.Gr.) provided resources for this study.
Conflict of interest
The authors declare that they have no conflict of interest