Rhamnolipids amendment improves soil properties and enhances microecological functions in the saline-alkali soil
Article information
Abstract
A laboratory-scale study was conducted to investigate the effect of rhamnolipids (RLs) supplement on the amelioration of saline-alkali soils. The RLs supplement improved the soil aggregates stability and promoted the formation of macro-aggregates which increased by up to 10.84% and 15.92% in alkaline soil and saline soil, respectively. In addition, RLs amendment led to a pH reduction from initial 8.87 to 7.80–7.84 in alkaline soil, and a salt rejection up to 20.72% in saline soil, remarkably alleviating the saline-alkali stress on microorganisms and plants. Meanwhile, microbial growth and activity as well as the seed germination performance were greatly improved in both types of soil. Furthermore, RLs addition greatly altered the microbial community structure and supported the proliferation of bacterial species (e.g., Pseudomonas oleovorans, Pseudomonas stutzeri, and Alcanivorax dieselolei) that favored the improvement of soil properties and nutrients circulation, thus markedly enhancing the microecological functions including carbon and nitrogen metabolisms. Further supplement of γ-PGA only exhibited promoting effect on aggregates formation and microbial growth and activity. The findings obtained in this study prove the application of RLs as a promising approach for saline-alkali soil amelioration.
Abstract
Graphical Abstract
1. Introduction
Soil salinization and alkalization are global concerns due to their restriction on land use efficiencies and agriculture production [1]. It is estimated that approximately 7% of the land area is influenced by high salinity to varying degrees, and the saline-alkali soil area is still increasing at a rate of 10% yearly [1, 2]. Xinjiang Uyghur Autonomous Region, one of the major agricultural provinces in China, is suffering from severe soil salinization, with nearly 32.07% of the total area of arable land remaining uncultivable due to the high salinity and water shortage [3]. The arid climate and severe evapotranspiration are the main reasons for the soil salinization in Xinjiang [4, 5]. Traditional approaches for saline-alkali soils amelioration including irrigation and drainage, sand mixing and mulching, and chemical amendments have been restrained due to the low cost-effectiveness, potential secondary pollution, and low efficiencies [6]. Therefore, developing a cost-effective and eco-friendly approach is vital to mitigate the severe soil salinization and alkalization.
Rhamnolipid (RL) is a type of glycolipid metabolites derived from microorganisms, possessing both hydrophilic and hydrophobic domains [7]. It has a variety of merits, such as low toxicity, good biodegradability, and high environmental compatibility [8, 9]. Due to the presence of hydrophilic groups, the addition of RLs to saline-alkali soil could be beneficial in reducing the water evaporation in the soil through the combination of water molecular and hydrophilic domains. Besides, previous research suggested that RLs can improve soil’s water conservation capacity by increasing the soil’s surface wettability and enhancing water transportation [10]. In addition, RLs can chelate a wide range of metal ions and promote their leaching and removal, implying the potential role of RLs in ameliorating salinized soil [11]. Meanwhile, Wen et al. [12] reported that RLs supplement can enhance the metal ions absorption by plant roots through the formation of lipophilic complexes, demonstrating that RLs can help to alleviate the salts accumulation in soils. Moreover, under alkaline conditions, H+ will be ionized from RL molecules, which further neutralize the hydroxide radicals in alkalized soil, thereby reducing its pH [13]. Although RLs exhibit great potentials for ameliorating saline-alkali soils, so far, very little attention has been paid in this regard.
Poly-γ-glutamic acid (γ-PGA) is a biosynthetic and environmental-friendly fertilizer with biodegradable and biocompatible properties [14]. Accumulated evidence suggests that γ-PGA plays an important role in water and nutrient conservation in salinized soils [15]. To the best of our knowledge, the coupling effect of γ-PGA and RLs on saline-alkali soil amelioration has not previously been investigated.
Microorganisms are important driving force for the circulation and transformation of soil substances [16]. Exploration of microbial community compositions during soil amelioration can provide deep insights on the underlying mechanism. Hence, the main objectives of this study were to 1) compare the effect of RLs supplement on the amelioration of saline soil and alkaline soil collected from Xinjiang; 2) explore the interactive effect of RLs and γ-PGA addition on saline-alkali soils; and 3) investigate the microbial community structure shift in response to the addition of RLs or γ-PGA and RLs. This study will provide insightful information for developing effective saline-alkali soils amelioration strategies.
2. Materials and Methods
2.1. Chemical Reagents
RLs used in this study was fermented by a bacterial strain, Pseudomonas aeruginosa CH1. P. aeruginosa is a well-known RLs-producing species, and the strong ability in RLs production of this strain has been verified in a previous study [7]. The critical micelle concentration of RL is 150 mg/L. Other reagents including γ-PGA, fluorescein diacetate, nutrient broth, mineral salts, and nutrient agar were of analytical grade and obtained from Aladdin Industrial Co. (Shanghai, China).
2.2. Collection of Saline and Alkaline Soils
Both saline and alkaline soil samples were collected from Alar, Xinjiang Uyghur Autonomous Region, China (40.32° N,81.17° E, 1,014 m altitude) on Sept. 19th, 2019. The soil samples were randomly selected from the topsoil (0–20 cm). After removing the impurities (i.e., plant branches and stone), the soil was air dried, sieved through a 2 mm mesh, and thoroughly homogenized. The basic properties of the saline and alkaline soils are presented in Table S1.
2.3. Experimental Design
To investigate the effect of RLs addition on the amelioration of saline-alkali soil, a pot experiment was conducted with a randomized block design consisting of three main blocks, each containing five sub-blocks designated as C, 3R, 6R, 3RP, and 6RP, respectively. The diameter and height of cylindrical plastic pots were 12 cm and 12 cm, respectively. Each pot was filled with 450 g of soil. The pot experiment was performed by supplementing: 1) 3wt% sterilized water (C); 2) 3wt% RL solution (5 g/L) (3R), resulting in a final RL concentration of 150 mg/kg soil; 3) 3wt% RL solution (10 g/L) (6R), resulting in a final RL concentration of 300 mg/kg soil; 4) 3wt% solution comprising 5 g/L RL and 1 g/L γ-PGA (3RP); 5) 3wt% solution comprising 10 g/L RL and 1 g/L γ-PGA (6RP). Saline soil and alkaline soil were separately used for the pot experiment, resulting in a total of 10 treatments, which were named C-A, 3R-A, 6R-A, 3RP-A, and 6RP-A for alkaline soil, respectively, and C-S, 3R-S, 6R-S, 3RP-S, and 6RP-S for saline soil, respectively. The moisture of the saline soil and alkaline soil was maintained at approximately 14.0% and 5.0%, respectively, by regularly supplementing water. The pots were statically placed at 25°C indoors for 10 weeks. Soil sample (30 g) at a depth of 6 cm was collected from each pot on day 0, 14 (2-week), 28 (4-week), 49 (7-week), and 70 (10-week) for further analysis.
2.4. Measurements
2.4.1. Electrical conductivity (EC) and pH measurements
After amelioration, soil pH was determined using a pH meter (soil/water, 1:5), and the EC was measured using an HQd portable meter (HACH, USA). Salt rejection (SR) was calculated using the following formula:
where EC1 represents the initial EC of saline-alkali soil, while EC2 represents the EC of soil after amelioration.
2.4.2. Soil aggregate distribution analysis
At the end of amelioration, the soil aggregates were analyzed using dry aggregate separation technique as previously described by Okolo et al. [17] with slight modifications. Briefly, 200 g of air-dried soil was collected at the end of amelioration and placed on a nest of sieves (4.75, 1, 0.50, and 0.25 mm) fixed on a rotary shaker (120 rpm) with a sieving time of 2 min. After dry sieving, the visible aggregates were carefully collected from each sieve, and the corresponding size ranges of aggregates were > 4.75, 4.75–1, 1–0.5, 0.5–0.25, and < 0.25 mm, respectively. Subsequently, the weight of aggregate within each size range was determined, and the percentage ratio (Wi) of a certain size of aggregate was calculated using the following formula:
where Wdi is the weight of aggregates within different size ranges.
R0.25, which represents the content of aggregates of > 0.25 mm, is calculated using the following formula:
where Mr>0.25 represents the weight of aggregates of > 0.25 mm.
Mean weight diameter (MWD) was determined to evaluate the stability of soil aggregates and was calculated as follows:
where n is the number of aggregate ranges, X̄i is the mean diameter of aggregate within a certain size range, and Wi is the percentage ratio of a certain-sized aggregate.
2.4.3. Determination of soil microbial counts and activity
After amelioration, the soil microbial counts were determined using plate count method, based on a previously reported protocol [18]. Meanwhile, the fluorescein diacetate (FDA) hydrolase activity, which could reflect the microbial activity and growth [8], was measured as described by Schumacher et al. [19] with slight modifications. Briefly, 2 g sieved soil was added to a 50 mL flask containing 15 mL phosphate buffer solution (NaH2PO4 0.1 g/L, Na2HPO4 2.2 g/L, NaCl 8.5 g/L, pH = 7.6), followed by adding 0.2 mL FDA solution (1 g/L FDA in acetone) to the mixture to initiate the reaction. Subsequently, the flask was placed on a rotary shaker at 30°C and 100 rpm for 1 h. Then 15 mL of termination reagent (chloroform/methanol, 2: 1, v/v) was added to the mixture to stop the reaction. After centrifugation at 2,000 rpm for 2min, absorbance of the supernatant was determined at 490 nm. The FDA hydrolase activity was calculated using the following formula:
where A is the absorbance of supernatant at 490 nm, As is the absorbance of fluorescein standard solution at 490 nm, C is the concentration of fluorescein standard solution (2 μmol/mL), V is the volume of the total reaction system, W is the weight of soil sample, and T is the reaction time.
2.4.4. Determination of seed germination percentage and index
The seed germination experiment was conducted as previously described by Jain et al. [20] with modifications. Briefly, the aqueous extracts of soil before and after amelioration were prepared by shaking with sterilized water in a rotary shaker at 200 rpm for 1 h and filtering through Whatman no. 42 filter papers (2.5-μm pore size). The seeds germination test was carried out by placing 10 soybean seeds in a petri plate spread with filter paper. Subsequently, 5 mL of aqueous extracts was added to the plate, which was then covered with a blotting paper to prevent external interference. After that, the plate was incubated in an incubator at 25°C for 5 d and the number of germinated seeds and shoot length were recorded.
The germination percentage (GP) was calculated using the following formula:
The germination index (GI) was calculated as follows:
A control was conducted by replacing the aqueous extracts with distilled water, which achieved a GP of 100%.
2.5. PacBio Full-length 16S rRNA Gene Amplicon Sequencing
Total genomic DNA was extracted using an E.Z.N.A. Soil DNA Kit (Omega Bio-Tek, USA) according to the instructions. The concentration and purity of the extracted DNA were confirmed by a Nanodrop 2000 spectrophotometer (Thermo, USA). Polymerase chain reaction (PCR) was performed to amplify the full-length 16S rRNA genes using the protocol described by Shuai et al. [21]. Primers 27F (5′-AGRGTTTGATYNTGGCTCAG-3′) and 1492R (5′-TASGGH TACCTTGTTASGACTT-3′) were used for the amplification. The amplicons were pair-ended sequenced on the PacBio Sequel platform. The original sequence data were available online with an accession number of SRP325264. The raw reads were filtered by Cutadapt 1.9.1 and UCHIME v4.2 software to remove primers, adapters, and chimera. Subsequently, the clean sequences were processed with Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) and clustered into operational taxonomic units (OTUs) based on a 97% sequence identity using the UCLUST algorithm. The taxonomic classification was based on the National Center for Biotechnology Information (NCBI) Gene database. Alpha diversity indices including Chao1 richness estimator, abundance-based coverage estimator (ACE), Simpson, and Shannon were calculated using QIIME2. Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software were used to predict the functional genes potentially contained in the samples and arrange the genes into different metabolic pathways [22].
2.6. Statistical Analysis
In this study, all data were the mean value of three replicates. The significance of differences among different treatments were examined using a one-way analysis of variance (ANOVA) and the least significant difference (LSD) test by SPSS software (v19.0, IBM Corp., USA).
3. Results and Discussion
3.1. Changes in Saline-alkali Soil Properties
3.1.1. Soil aggregates distribution
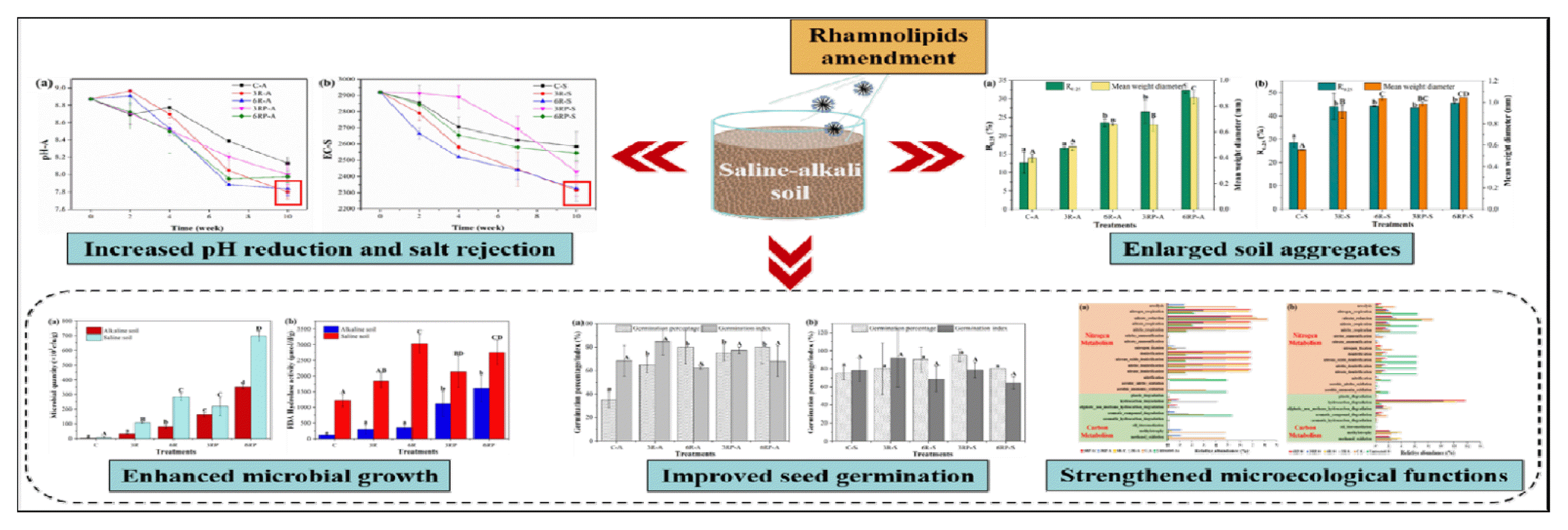
Soil aggregate size is an important parameter that reflects the stability of soil structure, and macro-aggregates generally exhibit higher stability and better water-holding capacity [17]. Initially, it was observed that high proportions of small-sized (< 0.25 mm) aggregates were presented in alkaline (92.57%) and saline soils (77.31%) (Fig. S1), suggesting its low stability and poor water-holding capacity. After RLs-mediated amelioration, the R0.25 and MWD, which are important parameters that reflect the aggregates size distribution and stability [15], were greatly improved (Fig. 1, Table S2). In alkaline soil, R0.25 and MWD in treatments supplemented with RLs exhibited 1.31–1.86 and 1.22–1.66 times higher values than the control treatment (R0.25 and MWD were 12.65% and 0.40 mm, respectively), respectively. Similarly, with respect to the saline soil, R0.25 and MWD in RLs-supplemented treatments exhibited 1.55–1.56 and 1.63–1.85 times higher than those of the control treatment (R0.25 and MWD were 28.38% and 0.56 mm, respectively), respectively. The results indicate that the RLs supplement greatly promoted the formation of macro-aggregates, suggesting the significant role of RLs in improving the soil aggregate stability and water-holding capacity. Besides, it was observed that the increase in RLs concentration or γ-PGA addition promoted the formation of macro-aggregates in alkaline soil. Although a similar pattern was shown in saline soil, the trend was not as clearly observable as that of the alkaline soil (Fig. 1b).
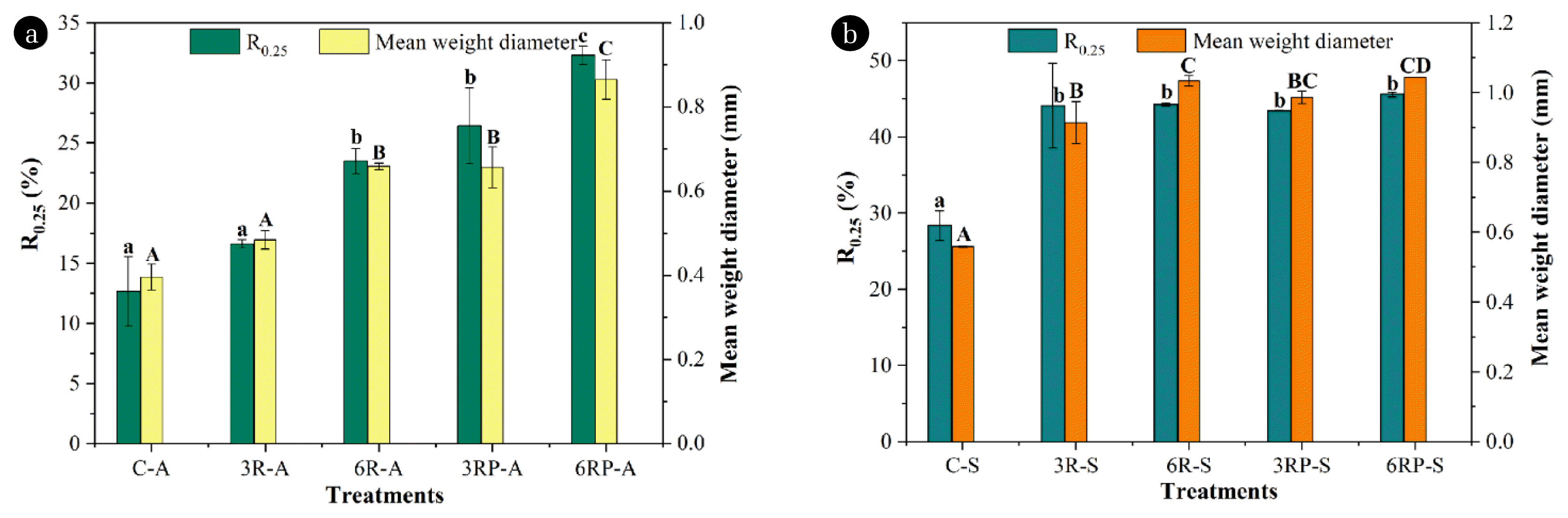
Effect of different treatments on the R0.25 and mean weight diameter in alkaline soil (a) and saline soil (b). R0.25 is the content of aggregates of > 0.25 mm; Mean weight diameter represents the stability of aggregates. The larger mean weight diameter, the higher stability of aggregates. The error bars represent the standard deviations.
In general, the formation of soil macro-aggregates is associated with the soil organic matter, and require the presence of substances that are able to cement the microaggregates [23]. In this study, the supplemented RLs and γ-PGA contain a large number of hydroxyl and amino groups on the molecular chain, with small particles and a large specific surface area, which would increase the organic matter content of soil and facilitate the cement of soil particles into macro-aggregates [15]. Additionally, the different amelioration patterns in both alkaline and saline soils could be possibly caused by the varied moisture contents in these two types of soil: the higher moisture in saline soil made it easier to form macro-aggregates, while in alkaline soil, the water shortage probably weakened the promoting effect of RLs and γ-PGA.
3.1.2. Soil pH and SR
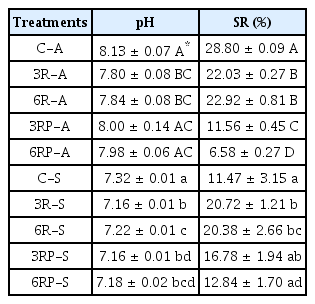
After 10 weeks of amelioration, soil pH and salinity were analyzed to evaluate the amelioration performance. As shown in Table 1, the pH decreased significantly (P<0.05) from an initial 8.87 to 8.13, 7.80, and 7.84 in C-A, 3R-A, and 6R-A treatments, respectively, suggesting the RLs addition promoted the pH reduction in alkaline soil, which primarily resulted from the neutralization of OH− by H+ ion from RL molecules [13]. However, the γ-PGA supplement weakened the promoting effect, leading to a slight increase in pH, compared to the treatments added with RLs only (Table 1). Likewise, the RLs supplement exhibited a good ability in reducing the alkalinity in saline soil, with the pH decreasing significantly (P<0.05) from 7.32 to 7.16–7.22. Similar findings have been reported by another study [24], in which biosurfactants application reduced the pH of forest soils.
In terms of the soil salinity, it decreased to varying degrees in different treatments (Table 1, Fig. S2). In alkaline soil, the SR were 28.80%, 22.03%, 22.92%, 11.56%, and 6.58% in C-A, 3R-A, 6R-A, 3RP-A, and 6RP-A treatments, respectively. The highest SR was shown in C-A treatment, gradually decreasing with the RLs and γ-PGA addition. During the formation of macro-aggregates, organic colloid and inorganic salts were conserved in these large particles, which probably led to the SR reductions in treatments supplemented with RLs and γ-PGA. With respect to the saline soil, the SR in C-S treatment was 11.47%, which increased by 9.25%, 8.91%, 5.31%, and 1.37% in 3R-S, 6R-S, 3RP-S, and 6RP-S, respectively, indicating that RLs and γ-PGA enhanced salts removal in the saline soil. Although the process of macro-aggregate formation may also conserve partial salts within the soil particles, the good salt removal performance mediated by RLs still led to a high SR in the saline soil.
3.2. Determination of Microbial Growth and Activity
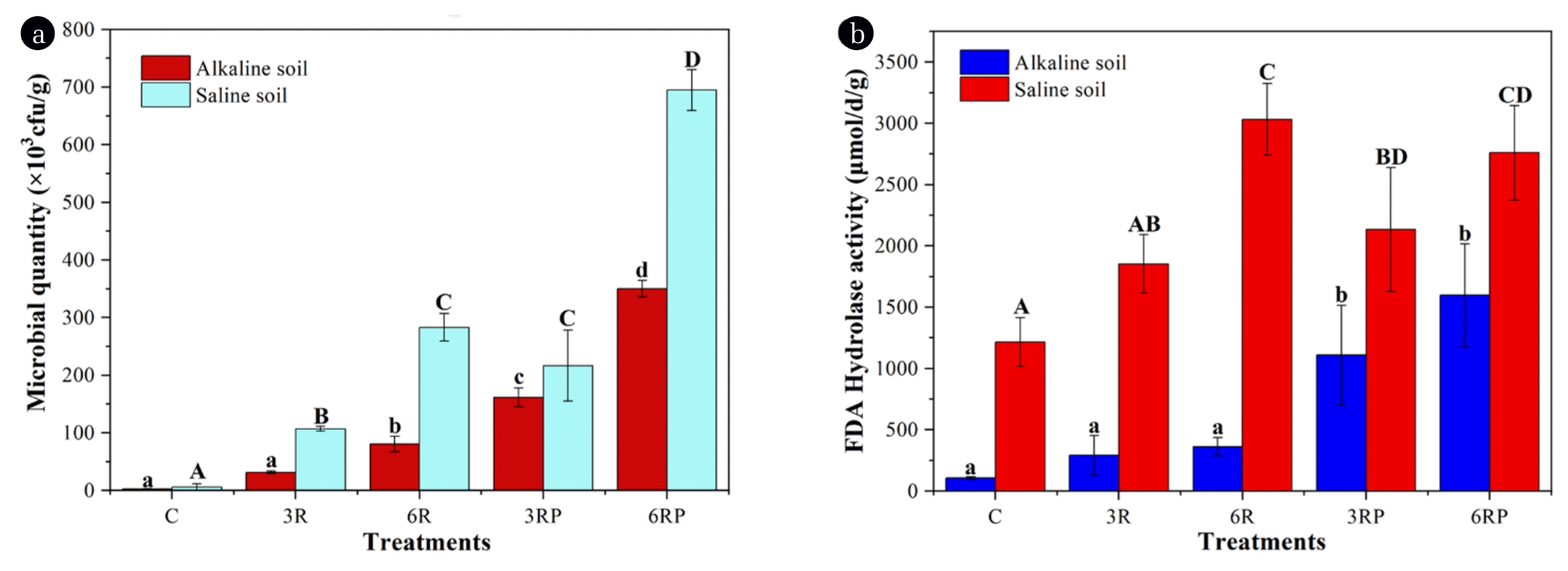
Microorganisms are important parts of soil as they participate in the processes of decomposition, energy flow, and nutrient cycle [25]. To investigate the effect of RLs supplement on the soil micro-organisms, microbial growth and activity in different treatments were determined, and the results are presented in Fig. 2. RLs supplement greatly improved the microbial growth in both types of soil. The microbial quantities in RLs-supplemented treatments were 12.6–47.2 times higher than the control treatment (Fig. 2a), suggesting a good growth-promoting effect of RLs on soil microorganisms. Besides, the microbial biomass in saline soil was higher than in alkaline soil. With respect to the microbial activity, a similar pattern was shown in different treatments. FDA hydrolase activity in RLs-supplemented treatments exhibited 1.5–3.4 times higher values than the control treatment (Fig. 2b). In this study, RLs could act as chelators, making soil nutrients more available for microbes [24]. Meanwhile, RLs can be utilized by microorganisms as carbon sources, directly supporting their growth and activity [7]. Also, γ-PGA could be applied as both carbon and nitrogen sources, further increasing the microbial biomass [26]. Furthermore, the reduction in soil pH and salinity also mitigated the external stress on micro-organisms, leading to increased microbial growth and activity. The results reported here indicated that RLs and γ-PGA addition could enhance the microecological functions of soil.
3.3. Effect of RLs on the GP and GI of Seeds
The GP and GI of seeds are important parameters that reflect the soil quality [20], and were measured to evaluate the amelioration performance. As shown in Fig. 3, the GP was improved in all RLs-supplemented treatments. Specifically, a significant (P < 0.05) increase in GP was observed in alkaline soil, suggesting the RLs addition greatly alleviated the stress of alkali on seed germination, making it conducive to increase the crop yield. Also, previous studies suggested that RLs could facilitate germination by increasing the permeability of external wrapping tissues and promoting water absorption [27, 28]. A similar result was reported by Singh et al. [29], that lipopeptide biosurfactant supplement greatly increased the GP of crop seeds. Additionally, GI in 3R treatments was higher than that of 6R treatments, indicating that high concentration of RLs could restrict the growth of seeds. Previous literatures showed that high concentration of RLs could form micelles that may solubilize cell membranes by forming mixed micelles with cell membrane lipids, leading to the necrosis of cells [7, 30]. In this study, the high concentration of RLs in 6R treatments might pose toxicity to the seed cells, thereby inhibiting the seed growth. Moreover, it was observed that the γ-PGA addition did not show promoting effects on seed germination, which probably resulted from the adverse influence of γ-PGA supplement on soil salinity and pH (Table 1), thereby inhibiting seed germination.
3.4. RLs Application Affects Microbial Community Compositions and Diversity in Saline-alkali Soils
3.4.1. Bacterial richness and diversity
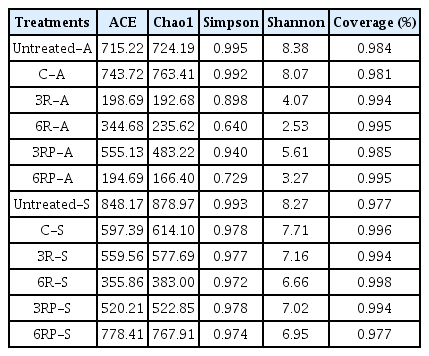
The full-length 16S rRNA gene sequencing was performed to investigate the potential alterations in soil bacterial community structure and diversity with the RLs supplement. In all RLs-supplemented treatments, the bacterial richness and diversity decreased dramatically at the RL concentration used in this study (Table 2), suggesting a selective enrichment of microorganisms in the presence of RLs. A similar pattern of decrease in bacterial diversity after RLs addition has been reported by Akbari et al. [31]. Besides, an obviously lower bacterial diversity has been observed in treatments with higher RLs dose, demonstrating that the selective enrichment is dose-dependent, which has also been indicated in other literatures [31, 32].
3.4.2. Microbial community compositions
Fig. 4 shows the major taxonomic groups at phylum level in different treatments. Obvious community structure shifts were shown in untreated and ameliorated soils, while the γ-PGA addition did not significantly alter the microbial community as compared to the RLs-amended treatments. The most abundant phylum in untreated and control treatment was Proteobacteria (31.50%–46.11%), followed by Acidobacteria (4.12%–17.19%), Actinobacteria (3.45%–11.39%), and Gemmatimonadetes (4.00%–12.00%). After amelioration, the relative abundances of Acidobacteria, Actinobacteria, and Gemmatimonadetes decreased to 0–2.28%, 0.06%–2.64%, and 0–2.18%, respectively, while the proportion of Proteobacteria further increased to 64.95%–79.06%. A previous study suggested that microbial community structure correlated positively with soil aggregate size [33]. Proteobacteria is reported to be capable of producing polysaccharides which is in favor of stabilizing soil aggregates [34]. Besides, many members affiliated with Proteobacteria are recognized as copiotrophic bacteria that prefer labile carbon in nutrient-rich conditions. Macro-aggregates generally harbor higher soil organic carbon than micro-aggregates [35]. Therefore, the higher proportion of macro-aggregates in RLs-supplemented treatments probably led to a higher relative abundance of Proteobacteria. Additionally, the relative abundance of Actinobacteriota increased from 3.77%–5.05% to 5.44–18.32% with the RLs supplement in saline soil. Microorganisms belonging to this phylum are typically thought to be aerobic obligate organoheterotrophs [36]. The addition of RLs and γ-PGA probably led to the proliferation of this phylum. In addition, Actinobacteriota generally behaves like fungi in soil, decomposing the organic matter of dead organisms and producing nutrients that can be taken up by plants [37]. Therefore, the presence and increase in relative abundance of this phylum are conducive for raising the crop yield in saline-alkali soil.
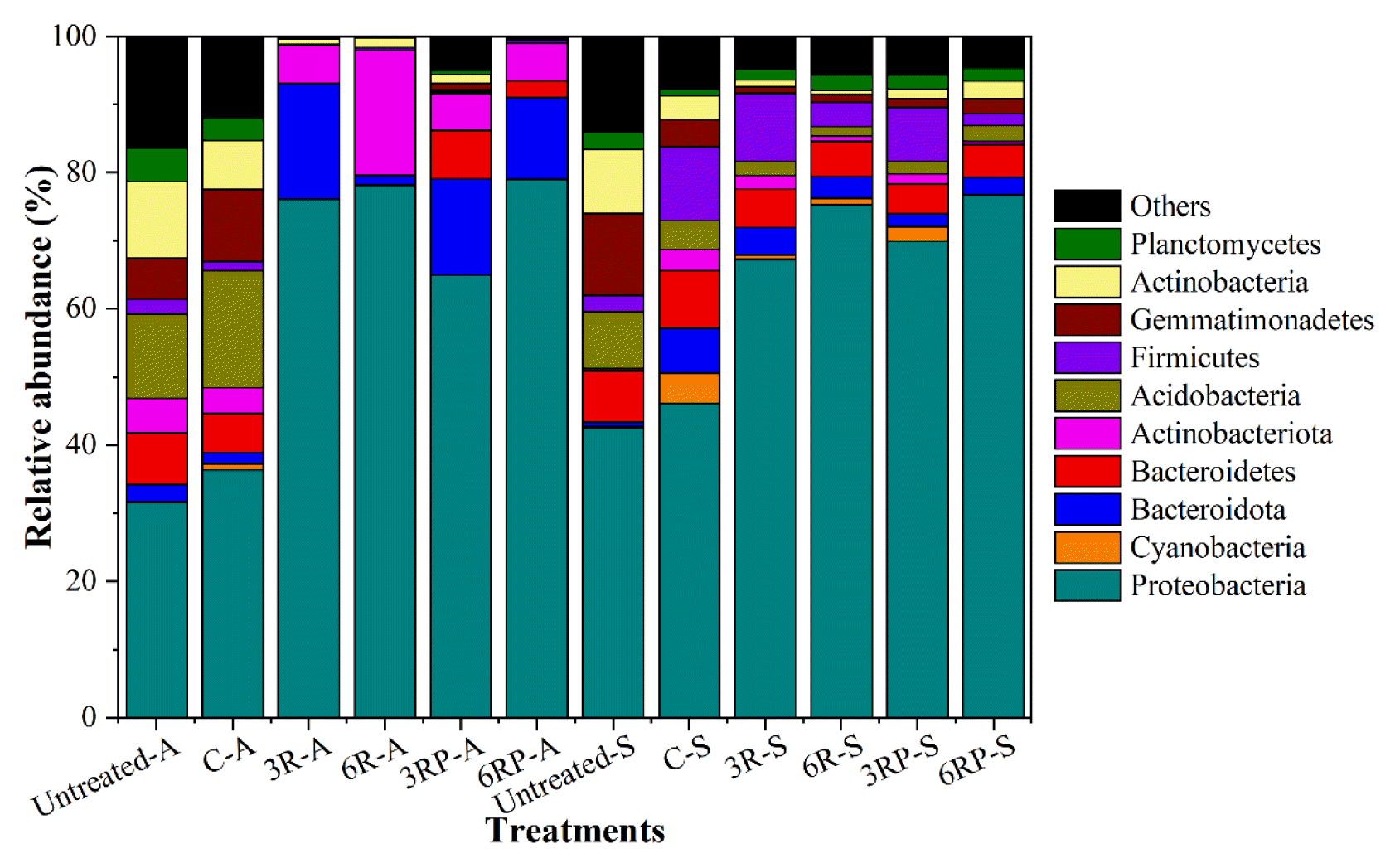
Effect of different treatments on the relative abundance of major taxonomic groups at phylum level in two types of soil. The most abundant operational taxonomic units (OTUs) at phylum level for all treatments are listed, and all other OTUs were combined and shown as “Others”.
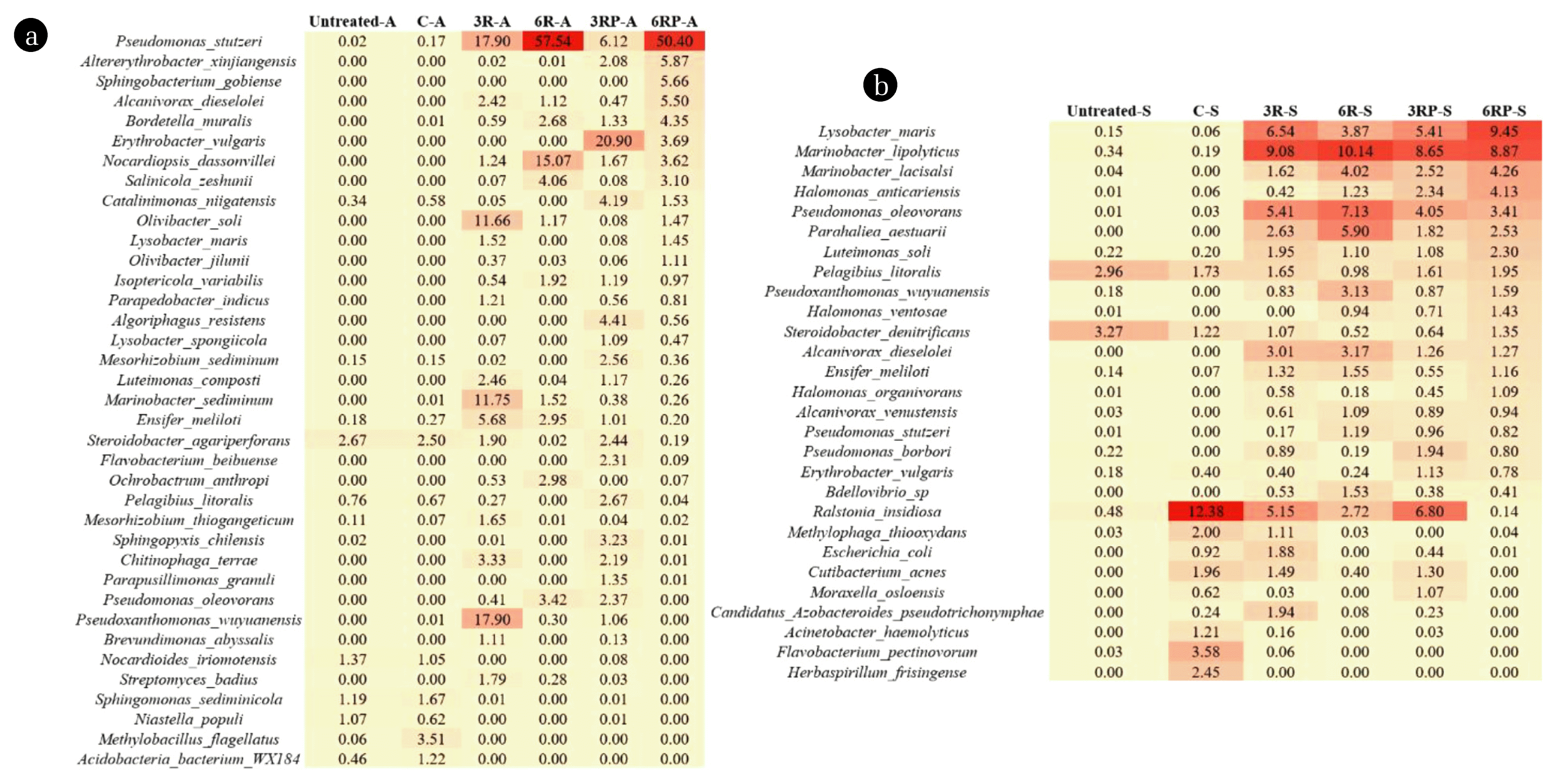
To obtain a better resolution of the microbial composition of the saline-alkali soil, a species-level identification was further carried out. As shown in Fig. 5, the abundances of major identified bacterial species in different treatments were presented. In alkaline soil, a sharp increase in relative abundance of Pseudomonas stutzeri from 0.17% (C-A treatment) to 6.12%–57.54% (RLs-supplemented treatments) was shown (Fig. 5a). Pseudomonas stutzeri can degrade a wide range of substrates and is adapted to growing in a weakly alkaline environment (pH 7.6–8.0) [38]. In this study, the supplement of RLs in alkaline soil led to a decrease in pH and provided a suitable environment for the growth of Pseudomonas stutzeri, thereby increasing the relative abundance of this species. Besides, a previous study showed that Pseudomonas stutzeri was capable of producing exopolysaccharide [18], which supports the stabilization of the soil aggregates. Other bacterial species including Alcanivorax dieselolei and Bordetella muralis, which have been reported to be affiliated with halophilic or basophilic micro-organisms [39, 40], also showed an increase in relative abundance, and were probably stimulated by the enhanced soil fertility and nutrient status. Concerning the saline soil, the microbial community shift was also observable, with Lysobacter maris (3.87%–9.45%), Marinobacter lipolyticus (8.65%–10.14%), Marinobacter lacisalsi (1.62%–4.26%), Halomonas anticariensis (0.42%–4.13%), Pseudomonas oleovorans (3.41%–7.13%), Parahaliea aestuarii (1.82%–5.90%), Luteimonas soli (1.08%–2.30%), and Alcanivorax dieselolei (1.26%–3.17%) exhibiting increases in relative abundance in RLs-supplemented treatments (Fig. 5b). These species have also been reported to thrive in saline environments [41–43]. RLs can release organic and inorganic nutrients from soil particles and promote the growth of these microorganisms [30]. Apart from that, RLs and γ-PGA can also serve as carbon and nitrogen sources for some microorganisms, leading to their proliferation. Moreover, some species such as Pseudomonas oleovorans, Pseudomonas stutzeri, Pseudomonas borbori, Alcanivorax dieselolei, and Alcanivorax venustensis are closely related to carbon and nitrogen metabolisms [44–46]. The increase in relative abundance of these species also demonstrated the enhancement of microecological functions of saline soil by the RLs addition.
3.5. Prediction of Functional Genes in Soil Samples with Different Treatments
PICRUSt analysis predicts the functional gene pathways in different samples, providing valuable insights into carbon and nitrogen metabolisms and how it shifts with the RLs amendment [47, 48]. As shown in Fig. 6a, the relative abundance of carbon-metabolism-related genes including methanol oxidation (0–1.09%), methylotrophy (0.005%–1.10%), and aromatic compound degradation (0.01%–1.73%) in RLs-amendment treatments dramatically decreased, while genes related to nitrogen metabolism including nitrate denitrification (2.92%–6.85%), nitrite denitrification (2.92%–6.85%), nitrous oxide denitrification (2.92%–6.85%), denitrification (2.92%–6.85%), nitrite respiration (2.93%–6.85%), nitrate respiration (2.99%–6.85%), and nitrogen respiration (2.99%–6.85%) showed higher relative abundance than those of the control treatment, implying that the RLs addition improved the nitrogen circulation in alkaline soil. Meanwhile, the increase in RLs concentration and γ-PGA supplement did not further promote the carbon or nitrogen metabolism (Fig. 6a). The promotion in nitrogen metabolism might result from the increase in the proportion of Pseudomonas stutzeri, which is a typical nitrifier and denitrifier, playing important roles in nitrogen cycles in soil [38, 46]. Regarding the saline soil, an opposite trend was exhibited in which RLs supplement promoted the carbon metabolism and relatively reduced the nitrogen metabolism (Fig. 6b). It was observed that the relative abundance of genes related to hydrocarbon degradation increased sharply. Previous studies showed that RLs addition can enrich hydrocarbon-degrading microorganisms, especially in oil-contaminated environments [31, 49]. In this study, the RLs could solubilize and release the hydrophobic compounds from soil surface, serving as a carbon source for microorganisms and leading to an enriched degrading population. Overall, the results suggested that the RLs supplement could be an effective approach in enhancing the microecological functions and nutrient circulation in saline-alkali soil.
Conclusions
This study reported on the RLs-mediated amelioration of saline-alkali soil. The supplementation of RLs promoted the formation of macro-aggregates and increased the aggregate stability. Besides, RLs addition led to a pH reduction in alkaline soil and a decrease of salinity in saline soil, which mitigated the stress of salts and alkali on microorganisms and plants, resulting in increased GP and GI. Meanwhile, RLs supplement greatly improved the microbial growth and activity, strengthened the microecological functions of the soil, and enhanced the nitrogen metabolism in alkaline soil while promoting the carbon metabolism in saline soil. The supplement of γ-PGA further promoted the formation of soil aggregates and improved microbial growth and activity. Moreover, results showed that the applied treatments were more effective for alkaline soil than saline soil. This study provides novel insights into saline-alkali soil amelioration and contributes significantly to the establishment of new strategies for mitigating the severe soil salinization and alkalinization.
Supplementary Information
Acknowledgements
This study was financially supported by the Zhoushan Science and Technology Department Project (2019C81056, 2020C41062), the Fundamental Research Funds for the Central Universities (2020QNA4045), and by the Research Funds of the Guangxi Key Laboratory of Theory and technology for Environmental Pollution Control (No.2001K004).
Notes
Conflict-of-Interest Statement
The authors declare that they have no conflict of interest.
Author contributions
S. L. (Postgraduate student) conducted all the experiments, data curation, supervision, and methodology. H. Z. (PhD student) wrote and revised the manuscript. C. C. (PhD student) conducted the formal analysis. F. Z. (Postgraduate student) conducted the formal analysis. G. Z. (PhD student) conducted the investigation. X. W. (Professor) conducted the investigation. C. Z. (Professor) conducted the validation, review, and project administration.