AbstractCoal ash, owing to its cost-effectiveness and greater adhesion with nitrifiers, has been used as a carrier for immobilizing nitrifiers when treating wastewater with a low C/N ratio and high nitrogen loading. Various environmental conditions significantly affect the nitrogen metabolic pathways of nitrifying coal ash. In this study, the nitrogen metabolic pathways of nitrifying coal ash propelled at different temperatures were investigated to evaluate the impact of seasonal variation on nitrifying coal ash. Results showed average NH4+-N effluent concentrations increased with temperature, and reached 3.37, 5.16, and 76.06 mg N/L with 1158 mg N/L influent under 15 °C, 25 °C, and 35 °C systems. The total nitrogen removal rate at a cooling temperature of 15 °C was higher than that at 25 °C and 35 °C. Further analysis showed Nitrosomonas and Nitrobacter were enhanced, whereas a significant evolution of autotrophic denitrifiers, Acidovorax, was enriched at 15 °C. An in-depth investigation of N-metabolism pathways suggested higher temperatures of 25 °C and 35 °C reduced the abundance of key genes involved in ammonia oxidation (amoABC, hao) and nitrite oxidation (narGH). Network analysis between N-metabolic functional genes and genera presented stronger relevance and richer sources between nitrifiers/denitrifiers and nitrification/denitrification-associated genes under cooler temperatures.
Graphical Abstract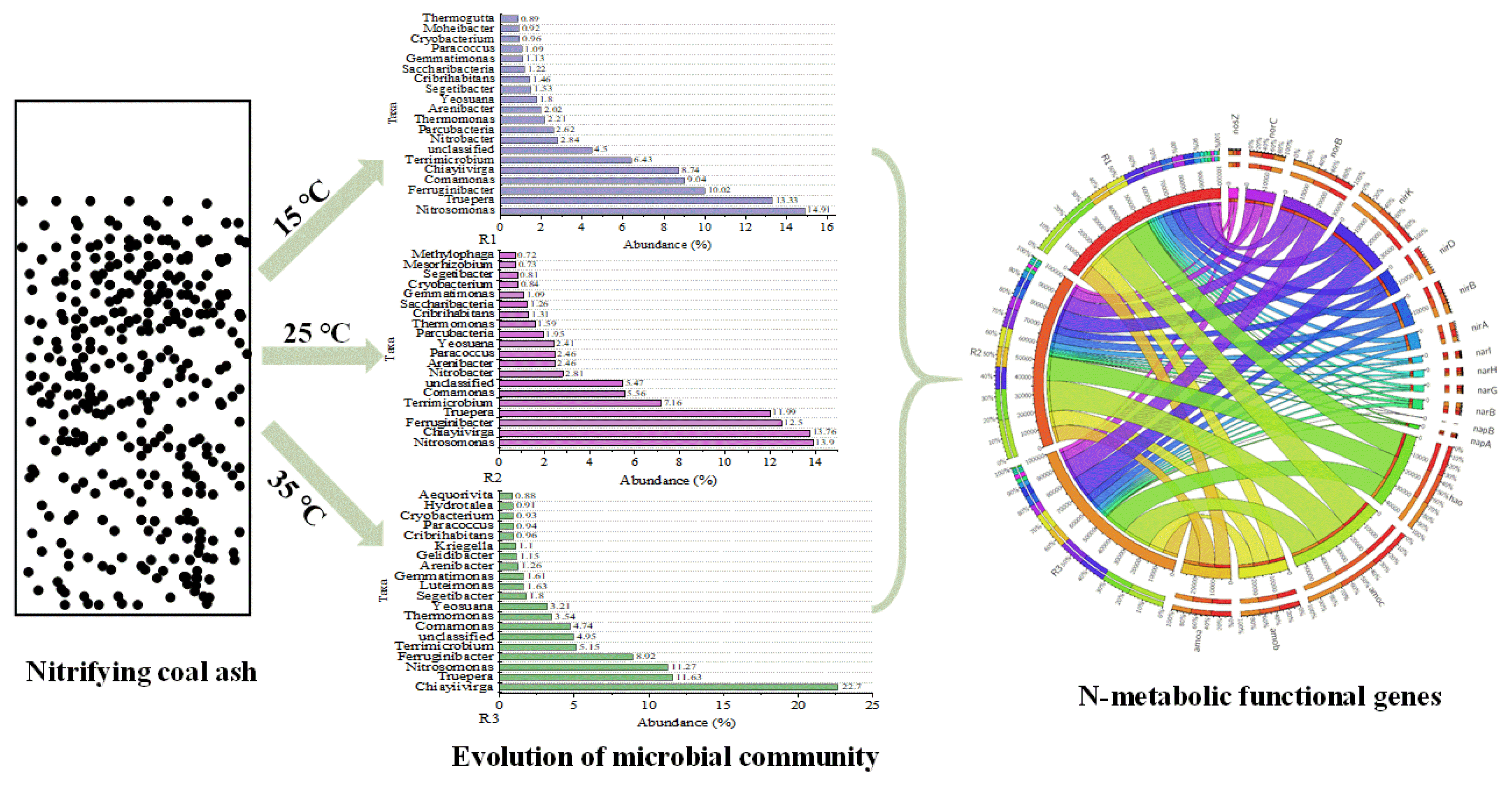
1. IntroductionThe activated sludge process has shown considerable advantages when treating wastewater with nitrogen [1], in which heterotrophic and autotrophic microbes work together. However, a limited C/N ratio inhibits the activity of heterotrophic microorganisms in multiple types of wastewater treatment [2], landfill leachate and municipal wastewater for example [3]. Previously, various improvement pathways aimed at efficient utilization of limited carbon sources have been developed from low-C/N wastewater, such as carbon addition, new strain development, and operating system update [4–6], which are restricted by high operating costs and management difficulties. Another effective way is to expand the role of autotrophic microbes in low-C/N wastewater treatment. Immobilization technology for autotrophic microorganisms is particularly common and enables greater economic efficiency [7, 8].
Immobilization technology fixes selected microorganisms on the carrier to maintain their high density and activity, which are generally obtained through physical and chemical methods [9]. Immobilization technology, such as anammox immobilization and nitrifier immobilization, has been successfully used to entrap microorganisms that are sensitive to environmental conditions and have slow growth [8, 10]. Combined entrapping for multiple types of nitrogen-metabolizing microorganisms can solve nitrogen pollution effectively. Choi et al. [11] used polyvinyl alcohol to entrap the ammonia-oxidizing bacteria (AOB) and anaerobic ammonia-oxidizing bacteria (AAOB), with a final NRR-C of 0.14 kg N/(m3 carrier· d) to treat synthetic wastewater with 315 mg/L NH4+-N. Li et al. [12] achieved the immobilization of nitrifying and denitrifying bacteria in construction waste, with the highest total nitrogen and NH4+-N removal efficiency of 91.97% and 78.82%, which improved the nitrogen removal performance. However, owing to the sensitivity of AOB and nitrite-oxidizing bacteria (NOB) to varying degrees of free ammonia (FA), which is advanced by ammonia concentration, temperature, and pH [13], the application of nitrifying bacteria received restrictions in this respect. Most of the existing studies have focused on low nitrogen loading systems [14–17], with little or no emphasis on high NH4+-N concentrations, let alone abundant nitrifying bacteria treating high NH4+-N wastewater. It is, therefore, crucial to investigate a novel method for immobilizing abundant nitrifying bacteria in treating high NH4+-N and low-C/N wastewater.
Coal ash, a particulate by-product of coal burning, is a promising material for potential applications in wastewater treatment [18]. It has been confirmed that coal ash can immobilize abundant autotrophic nitrifiers rapidly and the nitrifying coal ash is suitable for treating high concentrations of ammonia and low-C/N wastewater in previous studies [19, 20]. Most of the published studies on nitrifying coal ash focused on optimization of the cultivation conditions and analysis of the changes in the microbial community. However, when nitrifying coal ash is used for high N concentration and low-C/N wastewater under different temperatures, which respond to seasonal variation, the N removal efficiency and microbial community structures’ behavior are unclear. Beyond that, the transformation mechanisms of the key enzyme activities, and the network between it and microorganisms need to be studied.
Focusing on the above issues, the main objectives of this study were to investigate the nitrogen removal performance of nitrifying coal ash and the development of the microbial community structures when treating high-ammonia and low-C/N wastewater propelled by different temperatures. Moreover, key enzyme activities and network analyses involved in nitrogen metabolic pathways were also analyzed, which will provide valuable information to better understand the transformation of N occurring in the nitrifying coal ash systems with seasonal variation.
2. Materials and Methods2.1. Experimental Setup and OperationThree structurally identical sequencing batch reactors (SBRs) were used in this study. All reactors were made of Plexiglas, with a working volume of 2 L. The bioreactors were equipped with a series of heating devices, electric stirring devices, air pumps, and microcomputer time-controlled switches for temperature control, mixing, air delivery in the aerobic stage, and control of operation cycles, respectively. The 12 h operation cycle consisted of five stages: inlet, aeration, sedimentation, discharge, and stagnation, in which 10 h of operation was performed for the aeration stage.
All reactors were operated in parallel for 20 days. In previous studies, coal ash was taken from a power plant in Shenyang, China, and then used for the enrichment of nitrifiers by gradually increasing the NH4+-N concentration after acid and alkali pre-treatment, in which the relative abundance of nitrifying bacteria reached over 50%, and the initial biomass concentration was 1600 mg protein/L [20]. Synthetic wastewater was listed in Table S1, and the influent concentration of NH4+-N averaged 1158 mg/L. Reactors 1# (R1), 2# (R2), and 3# (R3) were operated at a temperature of 15±2 °C, 25±2 °C, and 35±2 °C, respectively, and the reactor from which the sludge was taken operated at room temperature and acted as the blank control. Dissolved oxygen (DO) concentration was controlled at 3–4 mg/L in the aerobic stage, and pH was controlled at 7.5–8.0 by 1 M sodium bicarbonate.
2.2. Analysis MethodsSamples were taken twice daily at the end of the aeration stage at a distance of 10 cm from the liquid level and then filtered with a 0.45μm filter before testing. NH4+-N, NO2−-N, NO3−-N concentrations were determined by spectrophotometer (SHIMADZU, Japan) after pre-treatment according to the Standard Methods for the examination of water and wastewater (APHA, 2005). Portable meters were used for measuring DO, temperature, and pH (WTW, Germany).
2.3. High-throughput Sequencing and Functional AnalysisSludge samples from all bioreactors were collected on the last day of each operation temperature and then centrifuged at 6000 r/min for cryopreservation at −80 °C until microbial community analysis. Sequencing was finished on the Illumina Miseq platform at Sangon Biotech Co., Ltd. (Shanghai, China). The PCR amplifications were conducted with the general primer 341F and 806R in T100™ Thermal Cycler (Bio-Rad, USA), which amplified with the V3–V4 regions of the 16S rRNA. Afterward, all sequences were successively processed by quality control and briefly, adapters, barcodes, and primers; meanwhile, sequences less than 200 bp and containing ambiguous “N” were removed. As result, a total of 134496 clean reads were generated with an average of 44832 reads of each sample. And then the reads were assembled into contigs with an average length of 460 bp. Raw sequence data involved in this study have been uploaded to NCBI Sequence Read Archive (accession number: SRR15885077 to 15885079).
PICRUSt was used to predict N-transformation metabolism functional genes based on 16S rRNA gene data. All functional predictions were categorized using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. N-transformation metabolism functional genes were identified by searching for KO numbers in functional annotation results.
2.4. Statistical AnalysisThe analysis of significance in NH4+-N, NO2−-N, and NO3−-N concentrations, and nitrogen removal efficiency in different reactors, as well as N-transformation metabolism functional genes, were performed using the t-test in SPSS 23.0 (Chicago, USA). All test values are indicated as means.
3. Results and Discussion3.1. Nitrification PerformanceAt 15 °C and 25 °C, the nitrification performances were fairly stable with effluent NH4+-N concentrations of less than 3.37 mg/L and 5.16 mg/L respectively, signifying over 99.9% ammonia was oxidized in R1 and R2 (Fig. 1). Temperatures of 15 °C and 25 °C had no significant impact on NH4+-N removal efficiency, which was consistent with another biofilm system [21]. When the temperature was increased to 35 °C, the effluent NH4+-N concentration increased significantly and reached 88.40 mg/L on the first day. As shown in Fig. 1, the temperature of 15–25 °C had no negative effects on NH4+-N transformation compared to the blank control throughout the experiment (Fig. S1). However, at 35 °C, the average influent NH4+-N concentration increased to 76.06 mg/L, and the NH4+-N removal efficiency decreased by 6.41%. When oxygen was limited, DO concentration decrease as temperature increased resulting in a negative temperature impact on the NH4+-N removal efficiencies. As for NO2− and NO3−, the average NO3− effluent concentrations were 610.58 mg/L, 670.11 mg/L, and 674.40 mg/L, and caused the accumulation of NO2− with average concentrations of 437.67 mg/L, 485.02 mg/L, and 502.29 mg/L in R1, R2 and R3, respectively. Temperature changes in these bioreactors resulted in different degrees of NO2− accumulation, which correlated with the microbial community structure and oxygen transfer [22]. There was a certain amount of total nitrogen loss in R1; TNR reached 4.79%, which decreased to 2.02% and 0.35% in R2 and R3, spectively, suggesting that an autotrophic denitrification process existed in R1.
Compared to the AOR and NOR in R1 and R2, the average values of biomass AOR in R3 were 4.35% (p=0.015) and 6.57% (p=0.009) lower, respectively, while the NOR was 4.94% (p=0.005) and 0.36% (p=0.830) higher, respectively, as shown in Fig. 2. The significantly lower AOR in R3 indicates that the biomass cultivated in R1 and R2 had a greater capacity for nitrification. However, the significantly higher NOR in R3 could be related to nitrogen removal by denitrification in R1. During the experiment, no changes were made to the influent ammonia loads, and the enhanced nitrification and TNR capacity in R1 and R2 could have been due to the increase in the enzyme activities and abundance of nitrifiers/denitrifying bacteria caused by temperature change.
3.2. Biofilm Microbial Communities
Table S2 and Fig. 3 show the diversity indices, phyla, and top 20 genera of the samples at a 97% similarity level. The diversity indices ranged from 3.33 to 3.54 (Shannon), 435.48 to 704.72 (Chao1), 532.59 to 954.41 (ACE), and 0.06 to 0.08 (Simpson) under coverage >99.6% (Table S2). Based on the diversity indices, the R3 bioreactor had minimal microbial diversity and richness, implying that a higher temperature reduced the microbial diversity in the nitrifying coal ash system in the present study. To further explore the differences among the three groups, the unique and shared operational taxonomic units (OTUs) of all samples are presented in Fig. 3 A, wherein 173, 90, and 66 OTUs are unique to R1, R2, and R3, respectively, and 173 OTUs were shared by them. A higher unique OTUs extended the community richness of the reactor, which was consistent with the Chao1 and ACE indices in R1 (Table S2). As shown in Fig. 3 A, the number of shared OTUs differed among the reactors, and the total number of OTUs decreased with the increasing temperature. Different species of microorganisms show different sensitivity to temperature changes, which led to different microbial community compositions.
Fig. 3 B shows the composition of the microbial communities at the phyla level in the samples. Over 90% of phyla in R1 and R2 samples consisted of 5 groups, Proteobacteria (48.69% and 48.47%), Bacteroidetes (21.91% and 24.63%), Deinococcus-Thermus (13.33% and 11.99%), Verrucomicrobia (6.78% and 7.85%), and Parcubacteria (2.62% and 1.95%), with Proteobacteria (52.42%), Bacteroidetes (25.8%), Deinococcus-Thermus (11.63%), Verrucomicrobia (5.17%), and Gemmatimonadetes (1.61%) in the R3 sample. The relative abundance of absolutely dominant phyla, Proteobacteria, was over 45%, which was consistent with those found in domestic sewage, piggery and tanning treatment processes [23–25]. Meanwhile, Proteobacteria and Bacteroidetes have been reported as the most abundant phyla, containing over 50% of the total phyla in an anaerobic/anoxic/oxic wastewater treatment system [26]. Most of these two phyla are heterotrophic microorganisms and significant reservoirs of mobile antibiotic-resistance genes (ARGs) [27]. Their abundance meant that there were sufficient organics in systems, such as soluble microbial products, which may have played an important role in forming biofilm with attachment and adhesion on coal ash.
Fig. 3 C shows the top 20 genera in the sludge samples at the genus level. The three dominant phylotypes were different: Nitrosomonas (14.91%), Truepera (13.33%), and Ferruginibacter (10.02%) in reactor R1; Nitrosomonas (13.9%), Chiayiivirga (13.76%), and Ferruginibacter (12.5%) in reactor R2; and Chiayiivirga (22.7%), Truepera (11.63%) and Nitrosomonas (11.27%) in reactor R3. Nitrosomonas, guaranteeing the oxidization of ammonia in systems, is widely distributed in wastewater treatment plants, animal manure, and strongly eutrophic environments [28]. Previously, the optimal temperature for nitrification was reported to be approximately 30 °C [29]. A contrary conclusion was observed in the current study; the negative influences of higher temperatures on AOB were ascertained, which resulted in a decrement in AOR. Owing to this, there was a decrease in saturated DO content when the temperature increased in biofilm systems [30]. Therefore, the 3–4 mg/L DO concentration appears to be the limiting condition for nitrifiers and nitrification in this study. Chiayiivirga has been identified as a major genus in multiple wastewater treatment systems, with the ability to degrade organic matter [31], which may come from extracellular polymers and dead bacteria. Ferruginibacter was found to be tolerant to the inhibition effect of aniline and played an important role in pollutant reduction [32]. Truepera had a significant effect on changes in the abundance of ARGs in the system [33]. Nitrobacter, a key NOB, was the dominating genera with approximately 2.84% and 2.81% relative abundance in both R1 and R2 samples, respectively, where only 0.56% of which thrived in R3. The abundance of Nitrobacter played a crucial role in oxidizing nitrite to nitrate in R1 and R2, insuring the radical oxidation of ammonia to nitrate. The NOB content under different temperatures was also connected with oxygen transfer in systems. In addition, Acidovorax, an autotrophic denitrifier [34], was enriched in R1 (0.88%), compared to R2 (0.03%) and R3 (0.01%). The TNR in R1 was 4.79%, accounting for 13.65 times that obtained in R3. This indicates that autotrophic denitrifiers supported by cooling temperatures substantially contributed to the TNR in R1. Only inorganic carbon and ammonium were available in the influent, therefore, the nitrogen removals by autotrophic nitrifiers and denitrifiers were considered to be the primary reaction processes over heterotrophic reactions.
3.3. Impact of Temperature on Key Enzyme Activities of N-transformationData on microbial community composition may not clearly provide an analysis of the key enzymes and functional genes involved in N-transformation. The activities of key enzymes involved in nitrification, denitrification, and dissimilatory/assimilatory nitrate reduction were measured using the KEGG Enzyme/Orthology databases. As shown in Fig. 4A, the key enzymes that catalyze NH4+-N oxidation, including ammonia monooxygenase (encoded by amoABC) and hydroxylamine dehydrogenase (encoded by hao), showed a decreasing trend in R3 compared with R1 and R2. Specifically, amoC in R3 was significantly reduced by approximately 10% (Fig. 4B). This indicates that ammonia oxidation metabolism was disrupted under heat stress. This outcome seemed to be a response to the weakening of nitrifiers in R3. Nitrite oxidoreductase, encoded by narGH, was significantly inhibited by higher temperatures in R3. Furthermore, the t-test results showed that temperatures of 25 °C and 35 °C significantly decreased the abundance of genes related to amoABC, hao, and narGH at the level of 0.05 (p=0.0009 and p=0.013, respectively). This further indicated that higher temperatures reduced the abundance of key genes involved in ammonia oxidation and nitrite oxidation and negatively affected nitrification efficiency.
For the denitrification pathway, there was a clear trend of increasing nitrate reductase activity with decreasing temperature, which was consistent with NOB and TNR. In-depth exploration revealed that the abundance of two metabolic pathways encoded by narGHI and napAB in R1 increased by 18.68% (25.32%) and 4.47% (5.40%), respectively, relative to that detected in R2 (R3) (Fig. S2). Thus, the obvious impact of higher temperatures was mostly linked to narGHI and its coding products. However, three other key enzymes associated with denitrification, that is, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase (encoded by nirK, norBC, and nosZ), showed few differences and were even slightly reinforced in R2 and R3, suggesting the crucial role of nitrate reductase relative to other processes during denitrification. Nitrite reductase, encoded by nirK and not nirS, was detected in this study (Fig. S2). According to previous reports, the co-occurrence of nirS, norBC, and nosZ genes suggests a shared regulatory mechanism, which may constrain complete denitrification in nirS-type denitrifiers. In contrast, no such genomic linkage pattern was observed for nirK, indicating that nirK-type denitrifiers are more likely to perform incomplete denitrification [35]. The deficiency of nirS-type denitrifiers reasonably explains the observed nitrite accumulation in all reactors. The abundance of nitrous oxide reductase demonstrated that higher temperatures were more inclined to result in N2 as the end product of the denitrification process. Similar results have been observed in previous studies in which the relationships between N2 emissions and temperature were found [36, 37].
A temperature of 15 °C induced enhancement of dissimilatory nitrite reductase, which may be the potential driving force for the slight NH4+-N residual in R1. In addition, the proportions of assimilatory nitrate reductase and nitrite reductase declined and fluctuated in all samples. The detailed reasons for these are discussed from a microbial origin perspective in the network analysis that follows.
3.4. Network Analysis between Microbes and functional GenesTo showcase the primary holders of N-metabolic functional genes (N-MFGs) and further investigate the roles of microbes in N transformation, a co-occurrence network was used for exploring and visualizing the connections between N-MFGs and the dominant genera (top 10). The relevance degree, calculated by the proportion of each N-MFG carried by a certain genus, was used to express the contribution of each genus to the N-MFGs in the present study. A total of 29 (R1), 30 (R2), and 29 (R3) genera sharing 26 genera were linked to N-MFGs by 122, 129, and 120 linkages in R1, R2, and R3, respectively (Fig. 5).
For R1, among the known genera, 7 N-MFGs were assigned to Nitromonas and Comamonas. In particular, Nitromonas were associated with amoABC, hao, nirK, norB, and norC, which explained 100% and 61.31% of the origins of amoABC and hao, respectively, demonstrating their leadership role in ammonia oxidation. Comamonas carried hao, napAB, nirK, norB, and nirBD, with relevance degrees of 37.17%, 65.13%, 26.26%, 28.84%, and 57.51%, respectively. This means that Comamonas was the primary repository of napAB and nirBD, which confirmed that Comamonas played a crucial role in nitrate reduction and dissimilatory nitrate reduction processes and might also be involved in denitrification in R1. Other potential holders included Nitrobacter (narGHI, nirK, nirBD), Thermomonas (narGHI, nirK, norB), and Arenibacter (napAB, nirK, norBC, nosZ, and nirBD). Notably, Nitrobacter and Arenibacter, the genera with high abundance in R1 and R2, showed strong relationships with denitrification and dissimilatory nitrate reduction processes. For example, narGH and narI were mainly found in Nitrobacter, with relevance degrees of 29.74% and 32.09%, respectively, in R1, indicating that Nitrobacter primarily performed complete nitrification/denitrification and dissimilatory nitrate reduction.
Most shared key holders in the R1, R2, and R3 samples had similar connections (Fig. 5). For example, Nitromonas maintained strong connections with genes responsible for ammonia oxidation, whereas Comamonas remained the largest supplier of napAB and nirBD, demonstrating their similar roles in each reactor. However, the differential relevance degree between shared genera and N-MFGs should be noted. For instance, the relevance degree between Nitrobacter and narI decreased by 2.45% and 24.76% in R2 and R3, respectively. Instead, Methylophaga was connected to narI with stronger degrees (7.59% and 6.54% in R2 and R3, respectively).
Beyond these shared connections, there were also special nodes and links in each network, making it more complex. New connections between Methylocystis, amoABC, and hao were also present in the R2 network, verifying the expanded role of Methylocystis in nitrification. Several heterotrophic bacteria were present in the network, for example, Defluviimonas in R1, Ottowia and Sphaerobacter in R2, and Gulbenkiania in R3. These bacteria are related to multiple genes such as norC, nosZ, and nirB. This may have resulted from the different relative abundances of these microbes in each system.
4. ConclusionsThis study provides insights into nitrogen removal efficiency, microbial structure, and metabolic mechanisms driven by different temperatures in nitrifying coal ash systems. In this study, higher temperatures of 25 °C and 35 °C caused a certain degree of suppression of NH4+-N and TN removal. A cooling temperature of 15 °C promoted the abundance of AOB and NOB, and autotrophic denitrifiers were detected in the reactor through 16S rRNA analysis, which may have contributed to the NH4+-N and TN removal. An in-depth investigation of N-metabolism pathways suggested that temperatures of 25 °C and 35 °C reduced the abundance of key genes involved in ammonia oxidation (amoABC, hao) and nitrite oxidation (narGH). Network analysis showed that the origins of the key N-MFGs did not differ significantly among reactors, except for the genes and genera related to nitrifiers and denitrifiers under cooler temperatures.
AcknowledgmentsThe study was funded by The Research Start-up Funds of Jiangsu University of Technology (Grant No. KYY19016) and the Major Science and Technology Program for Water Pollution Control and Treatment of China (Grant No. 2013ZX07202-010).
NotesAuthor contributions F. L. (Ph.D.) conducted the experiments and wrote the original draft. X. Z. (Assistant Professor) supported and edited the manuscript. C. L. (Assistant Professor) revised writing the initial version of the manuscript. X. H. (Professor) provided guidance on the overall research activities. References1. Cao S, Zhou Y. New direction in biological nitrogen removal from industrial nitrate wastewater via anammox. Appl. Microbiol. Biotechnol. 2019;103:7459–7466.
https://doi.org/10.1007/s00253-019-10070-3
2. Zhu RL, Wang SY, Li J, et al. Effect of influent C/N ratio on nitrogen removal using PHB as electron donor in a post-denitritation SBR. J. Chem. Technol. Biot. 2013;88:1898–1905.
https://doi.org/10.1002/jctb.4047
3. Gao X, Zhang T, Wang B, et al. Advanced nitrogen removal of low C/N ratio sewage in an anaerobic/aerobic/anoxic process through enhanced post-endogenous denitrification. Chemosphere. 2020;252:126624.
https://doi.org/10.1016/j.chemosphere.2020.126624
4. Yu G, Peng H, Fu Y, et al. Enhanced nitrogen removal of low C/N wastewater in constructed wetlands with co-immobilizing solid carbon source and denitrifying bacteria. Bioresour. Technol. 2019;280:337–344.
https://doi.org/10.1016/j.biortech.2019.02.043
5. Li W, Liu Y, Wu L, Ren R, Lv Y. Enhanced nitrogen removal of low C/N wastewater using a novel microbial fuel cell (MFC) with Cupriavidus sp. S1 as a biocathode catalyst (BCS1). J. Chem. Technol. Biot. 2019;95(4)
https://doi.org/10.1002/jctb.6309
6. Gong B, Wang Y, Wang J, et al. Intensified nitrogen and phosphorus removal by embedding electrolysis in an anaerobic-anoxic-oxic reactor treating low carbon/nitrogen wastewater. Bioresour. Technol. 2018;256:562–565.
https://doi.org/10.1016/j.biortech.2018.02.014
7. Warmuth W, Wenzig E, Mersmann A. Selection of a support for immobilization of a microbial lipase for the hydrolysis of triglycerides. Bioprocess Eng. 1995;12:87–93.
8. Wang Y, Li B, Li Y, Chen X. Research progress on enhancing the performance of autotrophic nitrogen removal systems using microbial immobilization technology. Sci. Total Environ. 2021;774:145136.
https://doi.org/10.1016/j.scitotenv.2021.145136
9. Zhang Z, Fan Z, Zhang G, Qin L, Fang J. Application progress of microbial immobilization technology based on biomass materials. BioResources. 2021;16:8509–8524.
https://doi.org/10.15376/biores.16.4.Zhang
10. Wang J, Liang J, Ning D, Zhang T, Wang M. A review of biomass immobilization in anammox and partial nitrification/anammox systems: Advances, issues, and future perspectives. Sci. Total Environ. 2022;821:152792.
https://doi.org/10.1016/j.scitotenv.2021.152792
11. Choi D, Khan MH, Jung J. Crosslinking of PVA/alginate carriers by glutaraldehyde with improved mechanical strength and enhanced inhibition of deammonification sludge. Int. Biodeter Biodegr. 2019;145:104788.
https://doi.org/10.1016/j.ibiod.2019.104788
12. Li H, Zhang X, Zhang X, Zhang Z. Immobilization of nitrifying/denitrifying bacteria onto construction waste and the elimination of pollutants from stormwater runoff. Desalin. Water Treat. 2019;152:82–91.
https://doi.org/10.5004/dwt.2019.23901
13. Kinh CT, Ahn J, Suenaga T, et al. Free nitrous acid and pH determine the predominant ammonia-oxidizing bacteria and amount of N2O in a partial nitrifying reactor. Appl. Microbiol. Biotechnol. 2017;101:1673–1683.
https://doi.org/10.1007/s00253-016-7961-2
14. Wang W, Ding Y, Wang Y, et al. Intensified nitrogen removal in immobilized nitrifier enhanced constructed wetlands with external carbon addition. Bioresour. Technol. 2016;218:1261–1265.
https://doi.org/10.1016/j.biortech.2016.06.135
15. Wang W, Ding Y, Wang Y, et al. Treatment of rich ammonia nitrogen wastewater with polyvinyl alcohol immobilized nitrifier biofortified constructed wetlands. Ecol. Eng. 2016;94:7–11.
https://doi.org/10.1016/j.ecoleng.2016.05.078
16. Zhang B, Long B, Cheng Y, et al. Rapid domestication of autotrophic nitrifying granular sludge and its stability during long-term operation. Environ. Technol. 2021;42:2587–2598.
https://doi.org/10.1080/09593330.2019.1707881
17. Wang S, Yang H, Zhang F, et al. Analysis of rapid culture of high-efficiency nitrifying bacteria and immobilized filler application for the treatment of municipal wastewater. RSC Adv. 2020;10:19240–19246.
https://doi.org/10.1039/d0ra01498b
18. Mushtaq F, Zahid M, Bhatti IA, Nasir S, Hussain T. Possible applications of coal fly ash in wastewater treatment. J. Environ. Manage. 2019;240:27–46.
https://doi.org/10.1016/j.jenvman.2019.03.054
19. Liu F, Hu X, Zhao X, Guo H, Zhao Y. An Efficient Way for Nitrifying Bacteria Enrichment with Coal Ash: Nitrification and Microbial Community. Water Air Soil Poll. 2017;228:1–7.
https://doi.org/10.1007/s11270-017-3553-8
20. Liu F, Hu X, Zhao X, Gao Y. Effect of carrier particle size on enrichment and shift in nitrifier community behaviors for treating increased strength wastewater. Water Environ. Res. 2021;93:1959–1968.
https://doi.org/10.1002/wer.1567
21. Zhu S, Chen S. The impact of temperature on nitrification rate in fixed film biofilters. Aquac. Eng. 2002;26:221–237.
https://doi.org/10.1016/S0144-8609(02)00022-5
22. Vogelaar JCT, Klapwijk A, Lier JBV, Rulkens WH. Temperature effects on the oxygen transfer rate between 20 and 55 °C. Water Res. 2000;34:1037–1041.
23. Sanchez O, Ferrera I, Gonzalez JM, Mas J. Assessing bacterial diversity in a seawater-processing wastewater treatment plant by 454-pyrosequencing of the 16S rRNA and amoA genes. Microb. Biotechnol. 2013;6:435–442.
https://doi.org/10.1111/1751-7915.12052
24. Chen J, Tang YQ, Wu XL. Bacterial Community Shift in Two Sectors of a Tannery Plant and its Cr (VI) Removing Potential. Geomicrobiol. J. 2012;29:226–235.
https://doi.org/10.1080/01490451.2011.558562
25. Niu Z, Guo H, Zhou Y, Xia S. Unraveling membrane fouling in anoxic/oxic membrane bioreactors treating anaerobically digested piggery wastewater. J. Environ. Chem. Eng. 2021;9:104985.
https://doi.org/10.1016/j.jece.2020.104985
26. Tang G, Zheng X, Li X, et al. Variation of effluent organic matter (EfOM) during anaerobic/anoxic/oxic (A(2)O) wastewater treatment processes. Water Res. 2020;178:115830.
https://doi.org/10.1016/j.watres.2020.115830
27. Niestepski S, Harnisz M, Ciesielski S, Korzeniewska E, Osinska A. Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mater. 2020;394:122544.
https://doi.org/10.1016/j.jhazmat.2020.122544
28. Nakagawa T, Takahashi R. Nitrosomonas stercoris sp. nov., a Chemoautotrophic Ammonia-Oxidizing Bacterium Tolerant of High Ammonium Isolated from Composted Cattle Manure. Microbes Environ. 2015;30:221–227.
https://doi.org/10.1264/jsme2.ME15072
29. Cho KH, Kim JO, Kang S, et al. Achieving enhanced nitrification in communities of nitrifying bacteria in full-scale wastewater treatment plants via optimal temperature and pH. Sep. Purif. Technol. 2014;132:697–703.
https://doi.org/10.1016/j.seppur.2014.06.027
30. Zhu S, Chen S. The impact of temperature on nitrification rate in fixed film biofilters. Aquacult. Eng. 2002;26:221–237.
31. Rodriguez-Sanchez A, Leyva-Diaz JC, Gonzalez-Martinez A, Poyatos JM. Linkage of microbial kinetics and bacterial community structure of MBR and hybrid MBBR-MBR systems to treat salinity-amended urban wastewater. Biotechnol. Prog. 2017;33:1483–1495.
https://doi.org/10.1002/btpr.2513
32. Yang M, Ma F, Xie Y, Li L, Xue Y. The SBR start-up performing simultaneous removal of organics, nitrogen and phosphorus from aniline wastewater: Pollutant removal efficiency and microbial community succession. Env. Pollut. Bioavail. 2021;33:104–112.
https://doi.org/10.1080/26395940.2021.1924079
33. Li Houyu, Zheng Xiangqun, Cao Haoyu, et al. Reduction of antibiotic resistance genes under different conditions during composting process of aerobic combined with anaerobic. Bioresour. Technol. 2021;325:124710.
https://doi.org/10.1016/j.biortech.2021.124710
34. Huang W, Gong B, Wang Y, et al. Metagenomic analysis reveals enhanced nutrients removal from low C/N municipal wastewater in a pilot-scale modified AAO system coupling electrolysis. Water Res. 2020;173:115530.
https://doi.org/10.1016/j.watres.2020.115530
35. Helen D, Kim H, Tytgat B, Anne W. Highly diverse nirK genes comprise two major clades that harbour ammonium-producing denitrifiers. BMC Genomics. 2016;17:155.
https://doi.org/10.1186/s12864-016-2465-0
36. Bizimana F, Timilsina A, Dong W, et al. Effects of long-term nitrogen fertilization on N2O, N2 and their yield-scaled emissions in a temperate semi-arid agro-ecosystem. J. Soil Sediment. 2021;21:1659–1671.
https://doi.org/10.1007/s11368-021-02903-4
37. Lai TV, Denton MD. N2O and N2 emissions from denitrification respond differently to temperature and nitrogen supply. J. Soil Sediment. 2017;18:1548–1557.
https://doi.org/10.1007/s11368-017-1863-5
Fig. 1Effluent nitrogen compounds concentrations (a–c), NH4+-N and TNR removal rate (d) under different temperatures in reactors. 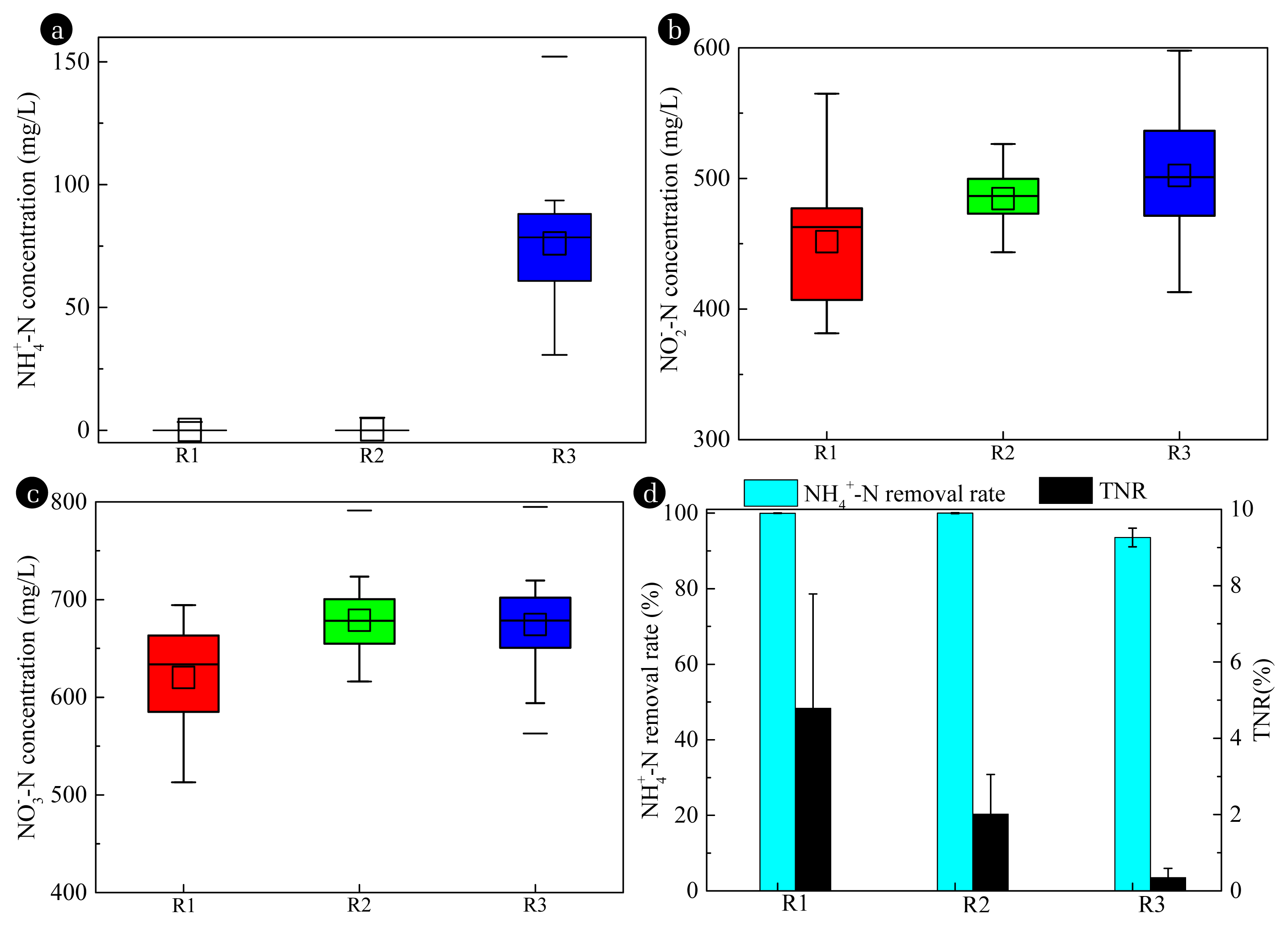
Fig. 3A) Venn diagram of the total number of OTUs, B) Top 5 phyla and C) Top 20 genera with the highest relative abundance of sludge samples. 
Fig. 5Network analysis of the co-occurrence between N-MFGs and top 10 dominant genera for A) R1, B) R2, and C) R3 reactors. Pink and other color nodes represent microorganisms and N-MFGs, and pink node size implies the number of connections. A connection indicates this genus is a potential hold of the N-MFGs, and the thickness of edges is proportional to the relevance degree. 
|
|
||||||||||||||||||||||||||||||||||||