1. Introduction
Nowadays industrial pollution is at an alarming stage due to increased anthropogenic activities. Textiles are among the basic needs of the human beings. The expansion of a textile industry contributes directly to the economic growth of any country [1]. Currently, the major anthropogenic source is textile industries. In textile industries, the main environmental concern is colored water, which originated from the dyeing process [2, 3]. Jodhpur and Balotra cities in western Rajasthan are the big clusters of textiles, dyeing and printing industries. Since dyes are formulated to be chemically and photolytically stable, they are highly persistent in natural environments [3–6]. They are chemically diverse in nature and can be divided into classes such as reactive, triphenylmethane, heterocyclic and polymeric dyes [3, 7]. Azo dyes are one of the most widely used dyes and can account for 70% of the total dye production [3, 4]. In the Jodhpur region, textile industries release untreated effluents through drainage channels into the Jojari River. While in Balotra, small scale industries generate a large quantity of wastewater which passes into the Luni River directly.
Serious concern about textile dyes and intermediate compounds was first raised due to its toxicity and carcinogenicity which can cause damage to the environment [8]. This highlights the need for treating textile dye-containing effluent before discharging it into water bodies [3, 7]. A wide range of biological, chemical and physical methods have been used to treat textile dye effluents [2, 9]. But as these processes are much expensive and generate amine residues containing sludge after degradation, regular consumption of such untreated toxic waters shows carcinogenesis in humans [1].
As an efficient alternative, biological treatment methods using aerobic or anaerobic microorganisms have received increasing interest owing to their high effectiveness and ecofriendly nature [9, 10]. Bioremediation is the microbial clean up approach, microbes can acclimatize themselves to toxic wastes and new resistant strains develop naturally, which can transform various toxic chemicals to less harmful forms [11]. Microbial processes can substantially contribute to efficient and reliable degradation of actual textile waste water [12]. Use of microorganisms for remediation purpose is thus a possible solution for environmental pollution since it includes sustainable remediation technologies. There are approximately thirty potential microorganisms investigated from different dye degradation experiments [13].
The capability of bacteria to metabolize azo dyes has been reported significantly [8]. Various bacterial decolorization studies investigated 100 percent efficiency using Bacillus sps. up to 90% [14], Pseudomonas sps. up to 70–100% [15, 16], E. coli up to 78.04% [15] and Klebsiella sps. up to 100% [17]. Bacillus strains are ubiquitous in activated sludge and have been found to degrade different dye groups [18].
The bacterial degradation of azo dyes produces aromatic amines which are carcinogenic and mutagenic [19]. Biodegradation of Congo red by bacillus thuringiensis supports the potential of other similar strains [20]. For successful bioremediation, contaminants should not accumulated as toxic or non-degradable intermediates [21]. Due to the different properties of azo dyes and their metabolites, such as solubility, volatility, and structure, analytical methodologies are needed. FT-IR spectrum enables the determination of both the type and strength of interactions that occur within azo dyes containing different functional groups after bacterial biodegradation, thus it acts as a valuable analytical tool.
In this study, we have isolated, screened and characterized the dye degrading bacteria collected from western Rajasthan province in India. Additionally, optimizations of various parameters were performed to compare the degradation efficiency of potential microbial isolates. Keeping this view to overcome the problems of partial degradation of textile dyes and the formation of toxic metabolites, a developed degrading bacterium from a dye contaminated environment was investigated for complete degradation of azo dyes and textile effluents without the formation of toxic aromatic amines. Furthermore, Fourier Transform Infrared spectroscopy (FT-IR) was carried out to evaluate the toxic effects of degraded products after treatment.
2. Materials and Methods
2.1. Soil and Waste Water Samples for Isolation Study
The textile water and soil samples were collected from different sites of the industrial area located near Jojari and Luni River in Jodhpur and Balotra of western Rajasthan, India for isolation of bacterial strains. As soil and water are the important sources for the isolation of microorganism. The particular soil conditions are enriched with wastewater. Soil samples were collected using the spatula and stored in a plastic bag and water samples were collected in clean plastic bottles and 60% nitric acid was added to preserve the components for further analysis. Samples were collected by standard procedure and stored at 4°C. The accurate location of the sampling site was recorded using the Global Position System (GPS) instrument. The sample sites at Jojari river is lying between 26°07′39.7″ north latitude to 72°59′05.9″ east longitude in Jodhpur district while the site at Luni river is located from 25°50′07.4″ north latitude to 72°12′30.7″E longitude in Barmer district of Rajasthan India. [22].
2.2. Dyes, Chemicals and Growth Media
All the dyes used in the present study were pure reactive azo dyes and collected from Ashoka dye chem. Ltd., Balotra. Red HE3B, Orange F2R, Yellow ME4GL, and Blue RC CT are commonly used commercial dyes, were selected for screening and decolorization experiments. All the dyes used in the study contain halogen and sodium sulfonate groups in their chemical formula. The chemicals used in this study were procured from Hi-Media India Ltd. All the experiments were carried out in triplicate and the resultant values were mean of those concordant sets.
The media used for growth, isolation and characterization studies was Nutrient Broth (Peptone: 5.0 g, Beef extract: 2.0 g, yeast extract: 1.0g/L, NaCl: 5.0 g/L, Agar: 15.0 g/L dissolved in1000 mL at pH 7.0) and Mineral salt media (Na2HPO4: 64g/L, KH2PO4: 15g/L, NaCl: 2.5g/L, NH4Cl: 5.0g/L, 1M MgSO4: 2 mL, 20% Glucose: 20 mL, 1M CaCl2: 0.1 mL dissolved in in 1,000 mL ) that were prepared as per standard protocols [9].
2.3. Isolation and Screening of Dye Degrading Bacteria
The bacterial strain used for azo dye degradation was isolated from water and soil from the industrial area located near Jojari and Luni River in Jodhpur and Balotra of western Rajasthan, India. Nutrient broth is the best medium used for the growth and isolation of dye decolorizing bacteria. Numerous colonies were obtained through serial dilution and streaking method. Each strain was then inoculated into the nutrient broth and incubated at 37°C on platform shaker Innova 2100, New Brunswick, Scientific) at 120 rpm for 24–48 h. A 10% (v/v) inoculum was transferred into a 250 mL Erlenmeyer flask containing 100 mL of Mineral Salt Medium (MSM) and incubated similarly. After 24 h, 10% (v/v) samples were sub-cultured into fresh MSM containing 10 mg/L of the respective dyes and further incubated as described above. Strains capable of utilizing fresh dyes as a nutrient source were plated on Nutrient Agar plates and incubated at 37°C for 24 h. Isolated colonies were taken and repeatedly streaked on nutrient agar to obtain pure cultures. The pure bacterial cultures were subsequently transferred into nutrient broth [23]. Each independent grown colony was named as SKNJ1, SKNJ2… SKNJ27.
2.4. Morphological and Biochemical Characterization of the Strain
Characterization studies were carried out based on morphological and biochemical methods. The morphological assay was done following standard method [24]. The cell suspension was spread onto a clean cover glass and was soaked in 3.5% glutaraldehyde for 6 h, and dried by treatment with 50, 70, 90, 95 and 100% ethanol, followed by overnight retention of the samples in a desiccator for the removal of moisture [25]. Biochemical tests were performed using the Hi media TM Identification Kit (Hi media India) according to the manufacturer’s instruction. The Biochemical tests included the detection of urease enzyme and sugar assimilation test. The positive tests were confirmed by color changes in the identification kit. The presence of oxidase was determined using Hi media DD 018 Oxidase discs (Hi media, India). Catalase activity was evaluated by transferring a loop of cells onto a microscope slide and adding a drop of 3% hydrogen peroxide solution [24].
2.5. Phylogenetic Characterization of the Strain
The pure cultures of isolated strains were analyzed by Yaazh Xenomics, India for sequencing of 16S rRNA gene (partial) and internal transcribed spacer (ITS) 1 and 2. The 16S rRNA gene is highly conserved and was used for the phylogenetic analysis of higher taxonomic levels, whereas the highly variable ITS region was used to differentiate the strain at lower taxonomic levels. The genomic DNA was extracted and the 16S rRNA gene and ITS regions were amplified by PCR using the following primers: 27F (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R (5′ AGAGTTTGATCMTGGCTCAG 3′). The obtained sequence was submitted to the NCBI GenBank database and a similarity search was carried out using the online BLAST program. The maximum likelihood was generated using 1000 bootstrap value with HKY85 employing PhyML 3.0 aLRT [26].
2.6. Optimization of Bacterial Isolates for Dye Decolorization
The bacteria selected from the initial screening were inoculated in Mineral salt media (100 mL) with respective dyes separately. The pH for bacterial growth was optimized in the range of 5–10 and different dye concentrations in the range 10mg/L, 20mg/L, 30mg/L, 50mg/L, 100mg/L, 200mg/L, 400mg/L, 600mg/L and 800mg/L was added. Similarly the bacterial inoculum was also serially added in the ascending range 5% (1 mL), 10% (2 mL), 15% (3 mL), 20% (4 mL) and 25% (5mL). Then all the cultures were incubated in a shaker cum incubator at 37°C and 120 rpm. The extent of decolorization was determined by measuring the absorbance at absorption maxima (λ max) of the samples at 24 h interval (i.e. 0, 6, 12, 18, 24, 48, 72 and 96 h). To ensure that all the decolorization was biologically mediated, MSM containing dye without inoculum served as the control was also carried out simultaneously [23].
2.7. Analytical Methods
2.7.1. UV-VIS spectrophotometric analysis
For analysis 2ml of the liquid sample was aseptically collected after the regular time interval and centrifuged at 7000 rpm for 15 min. The centrifuged cell-free supernatant samples were measured at λmax of 525 nm for Red HE3B, 590 nm for Orange F2R, 570 nm for Yellow ME4GL and 475 nm for Blue RC CT by Shimadzu double beam UV-VIS spectrophotometer [27]. The percentage of decolorization was calculated, according to Eq. (1):
Where, A0 = initial absorbance, At = absorbance after time t
2.7.2. FT-IR analysis
Biodegradation was characterized by FT-IR spectroscopy. The functional group characterization of the dyes before and after decolorization was studied. The samples prepared at the same concentrations were inoculated with the desired microbial inoculum to perform decolorization. These samples were centrifuged at 7000 rpm for 20 min. The degraded products present in culture supernatant were extracted using an equal volume of ethyl acetate. The liquid samples were directly analyzed by placing a drop on the thin-film cell. The FT-IR analysis of degraded products was carried out using PerkinElmer Spectrum Version 10.4.00 in the mid-infrared region of 400–4000 cm−1 with 16-scan speed [28]. IR spectrum was recorded.
3. Results and Discussion
Bioremediation of dyeing industry effluent by potential microorganisms has been proved to be an efficient tool. Important bacterial species including Bacillus, Pseudomonas, Enterobacter and Aeromonas have shown a tremendous capability to decolorize and detoxify a wide range of azo dyes [29].
3.1. Isolation and Characterization of Dye Degrading Bacterial Isolates
Dye degrading microbial strains were isolated from the sites nearby Jojari and Luni River. As the organic matter present in the effluent influence the concentration and variety of microorganisms in wastewater, therefore we also selected the organic matter rich wastewater in the present study. Besides, the wastewater effluent from the industrial area was thought to be rich in dye compounds; therefore the isolation of dye degrading microorganism was carried out. Twenty-seven microbial strains were successfully isolated from water and soil from these industrial areas to assess their ability to degrade azo dye. The isolated strains were preserved in agar slants at 4°C as well as in glycerol stocks at −20°C. These twenty-seven microbial strains were checked for dye decolorization assays. It was concluded from the dye decolorization experiments that only two strains SKNJ4 and SKNJ8 were found best to degrade dye concentration up to 800 mg L−1. Therefore, out of these twenty-seven microbial strains only above two strains have shown maximum decolorization percentage so the rest of twenty-five strains were not biochemically characterized. These two strains were preliminary identified as according to Bergey’s manual of systematic bacteriology and based on morphological and biochemical tests, as shown in Table 1. For confirmation of microbial strains, PCR and 16 S-rRNA sequencing was done. For phylogenetic characterization, the genomic DNA of bacterial strain SKNJ4 and SKNJ8 were extracted and the 16S rRNA gene and the ITS 1 (Internal Transcribed Spacer) and ITS 2 elements were amplified by PCR. The 16S rRNA gene is highly conserved and was used for the phylogenetic analysis of higher taxonomic levels, whereas the highly variable ITS region was used to differentiate the strain at lower taxonomic levels. The 16S rRNA gene and ITS region sequences (partial) of SKNJ4 and SKNJ8 strains were found to be 234 and 1000 bp respectively. The consensus sequence of the 16S rRNA gene was submitted to NCBI GenBank record (http://www.ncbi.nlm.nih.gov). Both strains were found to have more than 99% sequence similarity with Bacillus safensis strain NA-11 (Gene Accession Number: KC967061), Bacillus pumilis strain RHH76 (Gene Accession Number: HQ202554), Bacillus pumilis strain APS2 (Gene Accession Number: LT797528), Bacillus pumilis strain Dwi (Gene Accession Number: GQ131871), Bacillus sp. (Gene Accession Number: MF139392), Paenibacillus dendritiformis strain KP (Gene Accession Number: KX083535), P. dendritiformis strain PP (Gene Accession Number: KX082752), P. thiaminolyticus, strain RJ13 (Gene Accession Number: KR090600), respectively as shown in Fig. 1. The results for gelatin hydrolysis, casein hydrolysis, catalase test, starch hydrolysis, positive identification test profile for the dye degrading Bacillus species made it confirmed that isolated strains belong to the genus Bacillus species [30]. The phylogenetic approach to Bacillus taxonomy has been accomplished largely by the analysis of 16S rRNA molecules by oligonucleotide sequencing. This technique, of course, also reveals phylogenetic relationships. Some former members of the genus Bacillus were gathered into new families, including Acyclobacillaceae, Paenibacillaceae, and Planococcaceae, now on the level with Bacillaceae [31]. Therefore, the isolated bacteria were confirmed as Bacillus pumilis strain SKNJ4 (Gene accession number: MK670960) and Paenibacillus thiaminolyticus, strain SKNJ8 (Gene accession number: MK671362), respectively as shown in Fig. 1. A maximum-likelihood phylogenetic tree generated using PhyML 3.0 aLRT [26]. The program MUSCLE 3.7 was used for multiple alignments of sequences [32]. The resulting aligned sequences were cured using the program Gblocks 0.91b. This Gblocks eliminates poorly aligned positions and divergent regions [26].
In this study also, the selected strains were found similar degradation and dye decolorization kinetics as that of three different bacterial species, Bacillus species, Escherichia coli and Pseudomonas fluorescens collected from textile dye effluent [33].
3.2. Optimization Results
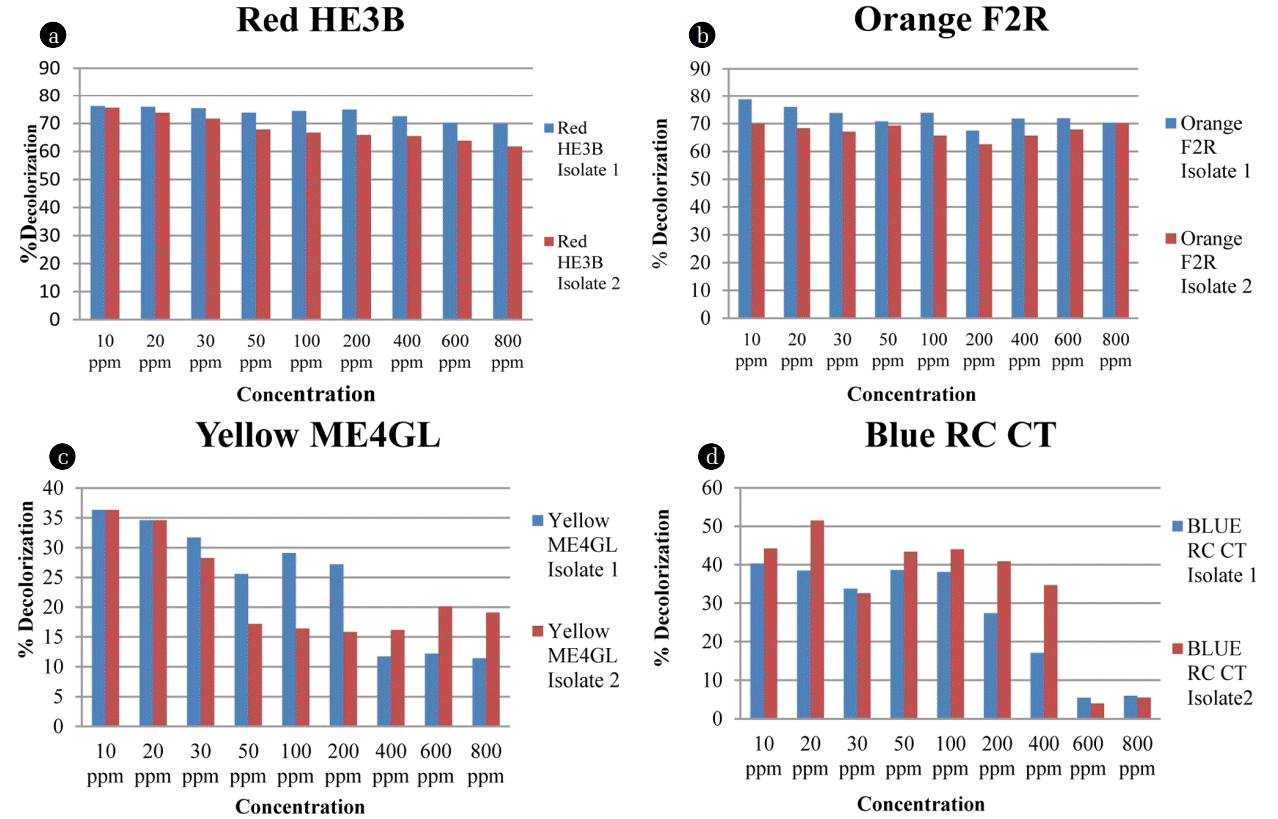
3.2.1. Effect of different dye concentration
The dye concentration of textile industry wastewater is normally in the range of 16–20 mg/L [7]. In this study effect of the higher initial concentration of dye (10–800 mg/L) on Bacillus species, decolorization potential was evaluated. Fig. 2 illustrates the decolorization percentage of experimental dye Red HE3B, Orange F2R, Yellow ME4GL and Blue RC CT with different concentrations by Bacillus pumilis and Paenibacillus thiaminolyticus. Both the species, at initial concentration of dye, efficiently decolorized Red HE3B (up to 76%) and Orange F2R (70–78%) followed by Blue RC CT (40–44%) and Yellow ME4GL (36%). Significant variation in the decolorization on increasing dye concentration by B. pumilis and P. thiaminolyticus is shown in Fig. 2. This variation suggested that both the isolates can be used to decolorize azo dyes. Higher decolorization percentage was investigated by many researchers [3, 10] with an enhanced incubation time using Aeromonas hydrophila. The variations of decolorization for different dyes by B. pumilis and P. thiaminolyticus might be significant due to the structural modification of the dyes. It has been reported that decolorization variation depends on the structure and complexity of dyes, particularly on the nature and position of substituents on the aromatic rings [3, 4]. The dyes used in this study, also undergo structural modifications in the presence of bacterial inoculum and other variables viz. pH, growth, incubation time monitored by FT-IR analysis.
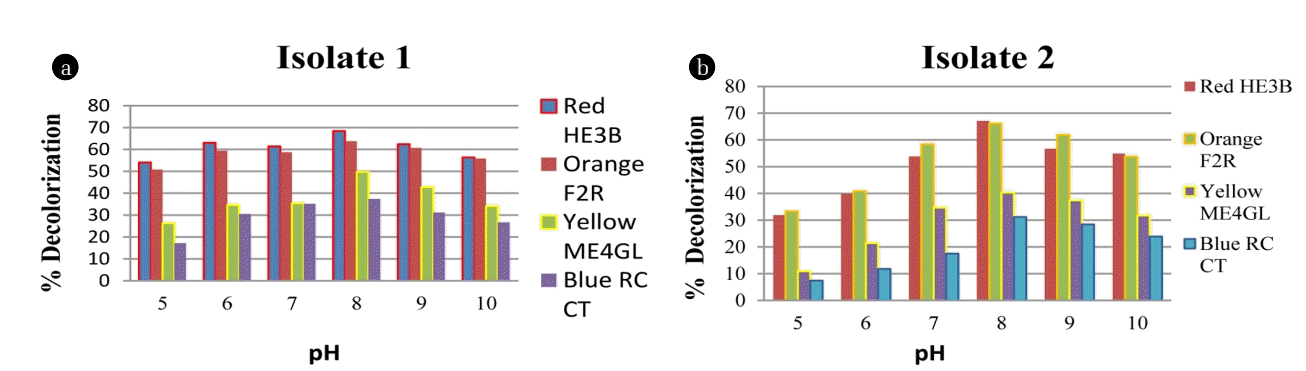
3.2.2. Effect of pH and inoculum size with time interval
The decolorization activity of both B. pumilis and P. thiaminolyticus was evaluated by adjusting the initial pH of the MSM from 5 to 10 is shown in Fig. 3. During the incubation periods, optimum decolorization results were obtained at pH 7 and 8. Fig. 3 illustrates the highest decolorization percentage of Red HE3B (68% and 67%) followed by Orange F2R (63% and 66%), Yellow ME4GL (49% and 40%) and Blue RC CT (37% and 31%) by B. pumilis and P. thiaminolyticus respectively. Both Bacillus sp. performs the best decolorization at neutral to alkaline pH range. Therefore these bacillus species are gaining importance for industrial wastewater treatment. During the dyeing process, textile industries are using different salt and chemicals that are found to be characterized by high salinity [3].
It was determined from various investigations and Fig. 4 also justify that in the present study, the decolorization percentage increases with increased inoculum size, at optimum pH range 8 within 24–72 h. Effect of pH was compared for the dye degrading strains Bacillus pumilis and Paenibacillus thiaminolyticus which displayed the highest decolorizing activity for Red HE3B (68.4 % and 67.1% respectively) at pH 8. Leena and Raj (2008) reported that Reactive black B was decolorized only 30% in 240 h, at neutral pH by Bacillus sp whereas Bacillus sp. N1 to N6 showed a 40–47% decolorization of seven dyes (mixed) in 336 h studied by Olukanni et al. [34–35]. The present study showed a high decolorization percentage as compared with previous findings. Previous research has illustrated excellent dye decolorization percent up to 48.15% decolorization with 10% inoculum concentration [36], while we find it up to 58% with 10% inoculum size. Dye decolorization percent increases up to 80% by both Bacillus species on increasing inoculum size as shown in Fig. 4. However, both the Bacillus species showed relatively better decolorization efficiency for Red (67%) and Orange (81.25%) dyes than Yellow (49.7%) and Blue (61.67%) dyes. Table 2 illustrates the bacterial decolorization percentage compared with previous findings.
3.2.3. Analysis of dye degrading product
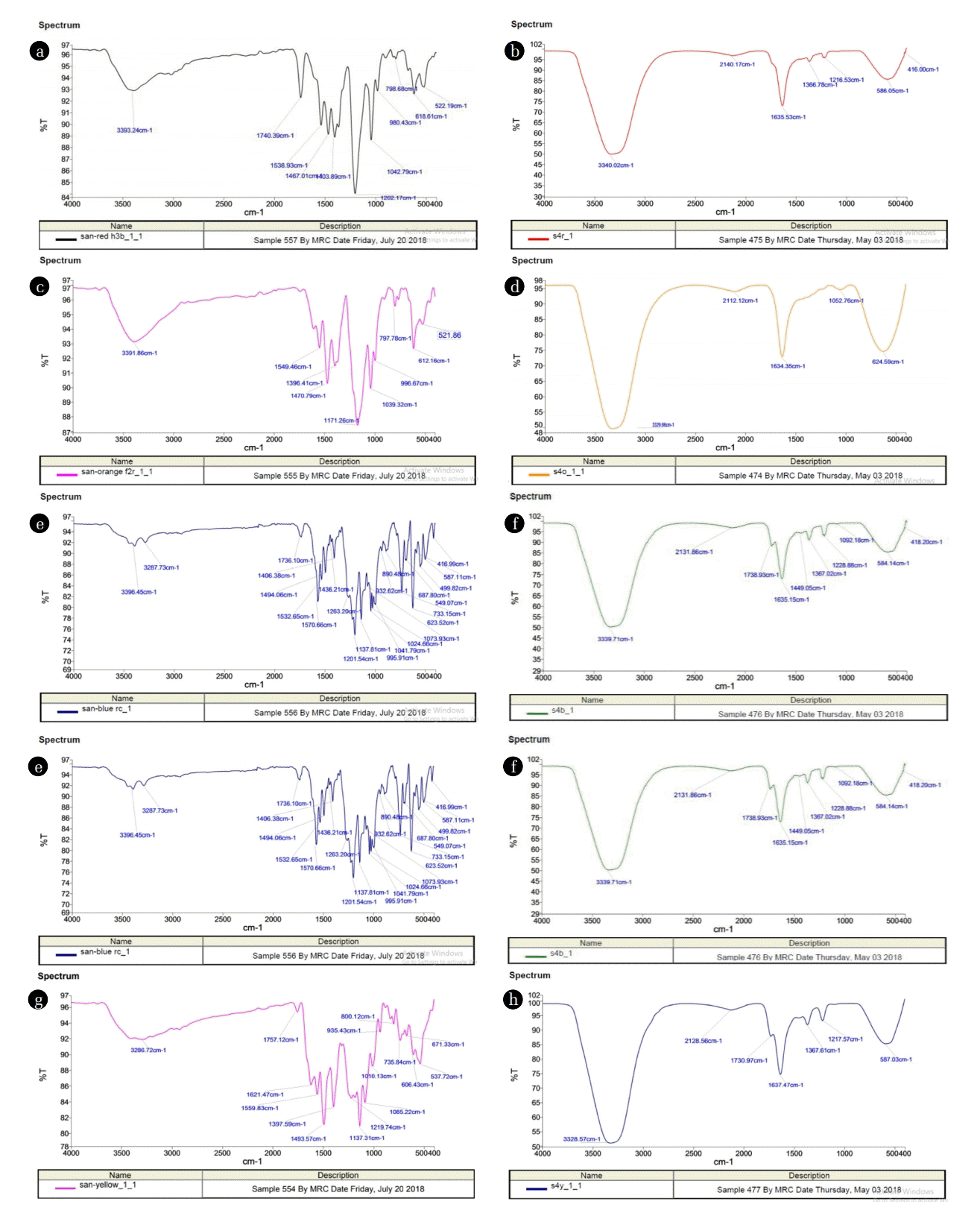
Decolorization studies of various dyes with UV-VIS spectroscopy confirmed the presence of spectral modifications. The FT-IR spectra of dyes showed prominent azo bond (N=N) vibrations which are identifiable between 1504 cm−1 and 1555 cm−1. The absorption frequency of sulfanilic acid is due to aromatic ring that occurs at 1573 cm−1 and 1498 cm−1 while C-N stretching occurs at 1240 cm−1. In the case of anthranilic acid, the aromatic ring absorption appears at 1574 cm−1 and 1453 cm−1 and that of C-N stretching at 1230 cm−1. The C-N stretching is observed at 1281 cm−1 while the aromatic ring band is next to the N-H band in the IR spectrum of aniline [39].
The FT-IR spectral results showed specific peaks corresponding to individual dye decolorization. Peaks in 3300–3500 cm−1 is normally observed for N-H and O-H stretching. In the FT-IR spectrum of parental dye named as Red HE3B, Orange F2R Yellow ME4GL, and Blue RC CT, the presence of -N=N- azo bond revealed at peak value between 1504–1595 cm−1. The peak value 1400 cm−1 showed the alkane C-H deformation and asymmetric stretching for the aromatic ring. The SOO asymmetric stretching at the peaks 1400–1025 cm−1 confirms the presence of sulphonic acid group in the parental dye, while CS stretching of sulphur-containing compound was depicted at 1000- 610 cm−1. These three peaks confirmed the presence of the SO3Na group in the dye structure. Peaks at 1250–1020 cm−1 represent the CN stretching that confirms amine compound in parental dyes.
The FT-IR spectrum of degraded products of Bacillus pumilis and Paenibacillus thiaminolyticus showed a significant change in positions of peaks when compared to control dye. There was an absence of peaks responsible for azo bond in degraded products obtained by both strains. Fig. 5 illustrates the spectra of the degraded products obtained from B. pumilis. The peak value at 3340.02 cm−1 (Red HE3B in Fig. 5(b), 3329.66 cm−1 (Orange F2R in Fig. 5(d), 3339.71 cm−1 (Blue RC CT in Fig. 5(f) and 3328.57 cm−1 (Yellow ME4GL in Fig. 5(h) indicated the −OH stretching, whereas peak at 1635.53 cm-1 (Red HE3B in Fig. 5(b), 1634.53 cm−1 (Orange F2R in Fig. 5(d), 1635.15 cm−1 (Blue RC CT in Fig. 5(f) and 1637.47 cm−1 (Yellow ME4GL in Fig. 5(h) represent the C=C stretching in conjugated alkene. Similar results were reported for Aeromonas sps. which revealed the peak value 1618.28 cm−1 and 1570.06 cm−1 for C=C stretching of the azo group in Acid Black- 24 dye [40] while in the case of Pseudomonas aeruginosa peak at 1653 cm−1 pointed toward the naphthalene product with C=O stretching for Ramazol black [41]. FTIR spectral analysis of acid blue 113 before and after decolorization by S.aureus and E.coli showed various peaks in the control dye which represents N-H stretching of amine or O-H and aromatic =C-H stretching at 3440 cm−1. The peaks at 1598–1566 cm−1 may be attributed to C=C bending, N=N stretching due to the azo bond was observed at peaks 1495 cm−1 and 1455 cm−1. The peak at 1190 cm−1 and 1102 cm−1 indicates SO2 symmetric and asymmetric stretching. Aromatic C-H out-of-plane bending vibrations at 816–627 cm−1 and in-plane bending vibrations at 1036 cm−1 were observed [42]. The FT-IR spectrum of degraded products obtained after decolorization by Paenibacillus thiaminolyticus as shown in Fig. 6.
The O-H intermolecular bonding and symmetrical stretching was indicated by peak value at 3400.42 cm−1 (Red HE3B in Fig. 6(b)), 3339.36 cm−1 (Orange F2R in Fig. 6(d)), and 3340.26 cm−1 (Yellow ME4GL in Fig. 6(h)). The peak obtained at 1584.35 cm−1 and 1635.57 cm−1 suggests COO stretching in aryl carboxylic acid in Red HE3B (Fig. 6(b)), Orange F2R (Fig. 6(d)), and Blue RC (Fig. 6(f)), respectively. The peak at 1642 cm−1 may be attributed to aryl carboxylic acid or quinone formed during decolorization by S. aureus [42]. The peak value reported at 1371.00 cm−1 (Red HE3B in Fig. 6(b), 1367.46 cm−1 (Orange F2R in Fig. 6(d)), 1366.29 cm−1 (Blue RC CT in Fig. 6(f)) and 1367.32 cm−1 (yellow ME4GLi in Fig. 6(h)) was responsible for SOO symmetric stretching suggesting that there may be an intermediate which was a sulfur-containing compound. A similar response at peak value 1339.21 cm−1 was shown by Aeromonas spp. for Orange 16 dye [28]. The spectrum of the dye degraded by E coli showed N-H amine stretch at 3382 cm−1, stretching of CH bond at 2196 cm−1 [42]. A considerable difference between the FT-IR spectrum of initial dye and metabolites obtained after decolorization confirmed the biodegradation of azo dyes.
4. Conclusions
The principal objective of this work was to evaluate the decolorization potential of selective microbial isolates which formed non-toxic metabolites developed from textile effluent and contaminated soils from Jodhpur and Balotra. Various factors affecting the decolorization process were investigated and used to optimize the decolorization process. The results of dye concentration conclude harmful effects with increased dye concentration, might be due to the toxic effect of biomass and inactivation of the enzyme responsible for dye degradation. The decolorization process increased with higher inoculum size, and decreased with increasing dye concentration while it showed higher potential within the pH range of 7–9 and within the time interval of 24–72 h.
After the microbial treatment of all dyes, a significant reduction in absorption was observed. A considerable difference between the FT-IR spectrum of parent dye and the metabolites obtained after complete decolorization by both isolates confirmed the biodegradation of parental dyes into lesser toxic metabolites. Thus, the present study may expose new heights in research in the areas of biological treatment of industrial effluents containing various dyes. The microorganism isolated and used in this study for dye degradation is a competent one and can further be exploited in industrial-scale application.