AbstractSynthetic wastewater was used to culture granular sludge for 21 weeks at three sequencing batch reactor activated sludge process (SBR) under different photoperiods. The growth of algae changed granular oxygen distribution, which affected the composition of microbial communities. Different photoperiods were able to alter the formation of granular sludge and the microbial community granules. In short photoperiod (12 h/d) sludge could generate symbiotic algae-bacteria granules in the SBR. By contrast, the long photoperiod (24 h/d) promotes the growth of algae and Ascomycota early, and then inhibits algae, especially chlorophyta (only 1.18%). In the end, algae-bacteria-fungi granules were formed in long photoperiod (LP). By GC-MS and model calculation, the lipid content of algae-bacteria granules was 33.71% more than that of pure aerobic granular sludge. However, the proportion of unsaturated fatty acids in algae-bacteria granules was as high as 43%, resulting in the quality of biodiesel prepared from algae-bacteria granule being slightly worse than that prepared from pure aerobic granular sludge. But both of them were better than biodiesel prepared from pure algae. The biodiesel content of algae-bacteria granule in short photoperiod (SP) and LP were 68.79 and 70.66 mg/g, respectively, which was better than that of pure aerobic particles (52.30 mg/g). Therefore, the formation of algae-bacteria granules is an effective way to remove nutrients and reduce harvesting costs.
1. IntroductionWith the expansion of the scale of domestic wastewater treatment, activated sludge, as a by-product, is also produced in large quantities, which puts huge burden on the environment. So, it is of great importance to find more sustainable and cost-effective ways that the by-product can be resource-based. The algal, as a low cost in-situ oxidant, have been used in the bacterial oxidation of activated sludge [1, 2], at the same time, abating the organic nutrients (nitrogen and phosphorous) and contamination sources via photosynthesis during wastewater treatment [3–5]. A large number of studies have shown that the excess sludge from algal-bacteria consortia system is considered as a potential source of biodiesel or fertilizer production [6, 7]. For instance, Liu et al. [8] found the maximum biodiesel yield of aerobic granular sludge (AGS) was 66.2 mg/g-SS, compared to conventional AGS biodiesel (35.4 mg/g-SS). In addition, the algal coupled process (algae-bacteria granular) could significantly improve the biodiesel yield of aerobic granules [9]. Therefore, algal-bacterial granule is becoming a promising biotechnology for wastewater treatment due to its excellent settling ability, high pollutant-removal efficiency, and simultaneous production of high-value output (biodiesel or fertilizer).
Up to the present, most of the studies on algal-bacterial AGS system mainly focuses on the generation of algal-bacterial AGS, operating conditions, and reactor settings, aiming at the stable and efficient performance on nutrients removal. For instance, Liu et al. [9] reported that algal community of granular consortia could be successfully formed by seeding microalgae, which have showed better nutrient removal ability and higher biodiesel yield. In addition, the effect of salinity has been studied on the culture and performance of algal bacterial AGS. Meng et al. [10] found that high salinity (1–3%) wastewater successfully cultivated mature (compact and smooth structure) algal-bacterial AGS and enhanced biomass growth, but had little impact on organics removal and the removal rate of total phosphorus was 57% (slightly lower). Moreover, algae could naturally grow and co-exist with AGS in a sequencing batch reactor (SBR) system exposed to natural sunlight, resulting in lower total nitrogen (TN) and total phosphorus (TP) removals [11, 12]. And few researchers reported that sunlight exposure promoted more rapid formation of algal–bacterial granules than aerobic granular sludge [11, 13]. Even though several studies reported the good photocatalytic performance of Engineering nanoparticles (NPs) driven by visible light [14–18], the environmental risk of NPs on biodiesel production from activated sludge has not been well studied. Clearly, previous attempts paid more attention to the effect of light intensity, salinity or NPs on nitrogen removal performance in conventional algal-bacterial symbiosis systems, while very few studies have addressed the impacts of illumination time on other nutrients removal, granular bioactivity, lipid production, biological (bacteria and algae) community and so on.
The goals of the current work were to (1) examine the ability and mechanism of N, P, and COD removal about algae-bacteria granular system; (2) quantitatively investigate the lipid content of algae-bacteria granules; and (3) reveal the composition of ecological community in the three SBRs.
2. Materials and Methods2.1. Experimental Set-up and OperationAlgae-bacteria granules were conducted in three identical sequencing batch reactors (SBRs) made of acrylic transparent plastic, each with an effective working volume of 3.2 L (D × H = 7 × 100 cm). All reactors were placed in an incubator which could avoid of their exposure to external light. These reactors were illuminated by cool white LED tubes (Opple Cool White-7W, 5700K) and the illumination intensity was 220 μmol/m2/s. Three different photoperiods were set in the reactor, which were 0 h/d, 12 h/d, and 24 h/d, namely dark period (dark), short photoperiod (SP), and long photoperiod (LP), respectively.
The reactors were operated sequentially in a 6-h cycle at room temperature (25 ± 2°C), consisting of 5 min feeding, 55 min of non-aeration, 278–296 min of aeration, 2–20 min of settling, and 2 min effluent discharge. During the aeration period, the air pump (AL-80, Arcadia, USA) provided mechanical aeration through the bubble diffuser at the bottom of each reactor, the inflow rate was kept at about 1.8 cm/s, and the dissolved oxygen (DO) was maintained at 7–9 mg/L. The effluent was discharged from a port at the middle of the reactor with a volume exchange ratio of 50%, resulting in a hydraulic retention time (HRT) of 12 h.
Each reactor was inoculated with 1.6 L sludge seeds, which were sampled from a municipal wastewater treatment plant in Tianjin, China. No external algae species was added into the seed sludge (aerobic sludge). The initial mixed liquor suspended solids (MLSS) concentration was 3.1 g/L with a sludge volume index (SVI30) of 110 mL/g and MLVSS/MLSS of at 0.79 in the three reactors. The compositions and average concentrations of the simulated wastewater were the same as our previous work [19], as shown: COD 600 mg/L, 50 mg NH4-N/L (NH4Cl), 10 mg PO4-P/L (KH2PO4), 10 mg Ca2+/L (CaCl2), 5 mg Mg2+/L (MgSO4·7H2O), 5 mg Fe2+/L (FeSO4·7H2O), and 1 ml/L of trace element solution [20, 21]. The pH in the reactors was maintained between 7.5–8.4 with sodium bicarbonate. The analytical grade purity of all these chemicals was supplied by Aladdin Industrial Corporation Ltd., China.
2.2. Lipid AnalysisLipids were obtained by adding 1.25 mL chloroform and 2.5 mL methanol (1:2, v/v) to each 100 mg of the dried, powdered biomass. The modified gravimetric method was used to measure the total lipid content of the granule samples [22]. The lipid composition of fatty acid methyl esters (FAMEs) was determined through the in situ transesterification method according to Liu et al. [8]. The FAMEs composition in the n-hexane phase was analyzed by gas chromatography (GC) in combination with tandem mass spectrometry (7890B GC/5977A MS, Agilent, USA) using Agilent HP-5MS capillary column (30 m × 250 μm × 0.25 μm).
2.3. Gas Chromatography-mass Spectrometry (GC-MS)In order to identify the biodiesel components of granule, the effluent was analyzed with GC-MS. The GC separation was performed on a HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm) (Agilent Technologies, CA, USA). Split ratio was 20:1 that was used at 280°C with helium as a carrier gas at 3 mL/min and the injection volume was 2 μL. The temperature program was as follows: 80°C, hold for 2 min, increase at a rate of 10°C/min to 280°C, and then hold for 2 min. A solvent delay time of 3 min was performed before the ion source was turned on and the mass spectra were acquired from m/z 30 to 550. The chromatographic peaks were identified using the NIST11 library (National Institute of Standards and Technology, Gaithersburg, MD, USA, http://www.nist.gov/srd/mslist.htm), and a match percentage was obtained by comparing the mass spectrum of a peak with that of a known compound from the library.
2.4. Biological Community AnalysisAlgae-bacteria granular consortia in the three SBRs was sampled at the end of the experiment (140 days) for bacterial and algal communities analysis. The detailed analytical method of microbial community and Eukaryote (include algal) community are as follows. For the identification and semiquantitative analysis of algae, an optical microscope (SDPTOP, CX40, China) equipped with a digital camera (CANON, 700D, Japan) was used. The algal species were identified according to “The Freshwater Algae of China Systematics, Taxonomy and Ecology” written by Hu Hongjun et al. [23].
2.4.1. Analysis of prokaryotic communityThe total DNA (100 μL) of granular sludge samples harvested from three SBRs were extracted by using Mo Bio PowerSoil DNA Isolation Kit (MoBio Laboratories, Inc., USA) according to the manufacturer’s protocol. After DNA extraction, polymerase chain reaction (PCR) and high-throughput sequencing were performed. Briefly, the rough full-length 16S rDNA gene was amplified by PCR with a forward primer V4. PCR mixtures (30 μL) contained 15 μL of Phusion Master Mix, 3 μL of each primer (two primer), 10 μL of diluted DNA and 2 μL of sterile distilled water. The sequences of the primers were as follows: primer 515F-GTGCCAGCMGCCGCGGTAA; primer 806R-GGACTACHVGGGTWTCTAAT. The PCR conditions were as follows: 95°C for 7 min, followed by 32 cycles at of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min and a final extension step at 72°C for 10 min. Then, the PCR products were purified with the Gene JET Gel Extraction Kit (Thermo Scientific). After quantification using the Qubit@2.0 Fluorometer (Thermo Scientific), the PCR products of all samples were taken for high-throughput sequencing on Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an IlluminaHiSeq2500 platform and 250 bp paired-end reads were generated.
2.4.2. Analysis of Eukaryotic (include algae) communityFor the granular sludge samples, the Mo Bio PowerSoil DNA Isolation Kit (MoBio Laboratories, Inc., USA) was used to extract genomic DNA. Illumina sequencing library preparation was performed according to best practices described by Ho et al. [24]. PCR mixtures (30 μL) contained 15 μL of Phusion Master Mix, 3μL of each primer (two primer), 10 μL of diluted DNA and 2 μL of sterile distilled water. The following PCR amplification targeted the V4 region of the 18S rRNA gene, and for this, the universal primers 528F (GCGGTAATTCCAGCTCCAA) and 706R (AATCCRAGAATTTCACCTCT) were used [24, 25]. Thermal cycling conditions were: 98°C for 2 min followed by 25 cycles of 98°C for 20 s, 63°C (16S), or 54°C (18S) annealing for 20 s, 72°C for 15 s, and a final extension step of 2 min at 72°C. Other analysis methods of eukaryotic community are the same as above.
2.5. Other AnalysesThe concentration of chlorophyll-a (Chl-a) and chlorophyll-b (Chl-b) were analyzed as described by Ritchie [26]. The dissolved oxygen (DO) concentration in the bulk liquor was measured with the same DO meter (HQ40d, HACH, USA). The morphology characteristics of the seed sludge, algae, and granules in reactors were observed using an optical microscope (SDPTOP, CX40, China) equipped with a digital camera (CANON, 700D, Japan).
3. Results and Discussion3.1. Formation of Algal-bacterial Granular Sludge at Different Illumination TimeAfter 2 weeks of operation, yellow particles with clear boundary are observed under microscope in three reactors, and gradually grew up (Fig. 1(a)). In SBR with SP and LP, light can cause algae growth and sludge particles turn green gradually (Fig. 1(a)). With regard to the source of algae, because it is an open system, the algae are inoculated from tap water or air. From week 3 to 21, the different growth trends of granular diameter and biomass were observed in SBR reactor, probably attributable to the rapid growth of algae in the SBR [19, 27]. After 1 month cultivation, white granules with average particle diameter of 0.52 mm were observed in all the reactors (Fig. S1(a)). The particle diameter increases with the increase of illumination time, and the main particle size was 1–2 mm in dark and SP, while about 50% of the particles in LP were larger than 4 mm after 5 months’ cultivation (Fig. S1(b)). MLVSS of all SBRs from 2.5 went to 9 g/L after 6 weeks’ cultivation, and tended to be stable in dark and SP (Fig. 1(b)). Long term light promoted the growth of algae in LP, which led to MLVSS reaching 12.5 g/L in 6 weeks, and then gradually decreasing to about 9 g/L. It is worth noting that Fig. 1(c) reveals that SVI dropped from the initial 110 to about 10 mL/g in three SBRs, and then slowly increased to about 20 mL/g. The higher biomass concentrations in SP and LP compared to the control (Drak) clearly support earlier findings with respect to that the MLVSS biomass increased accompanied with the illumination conditions [28, 29]. In the first three weeks, the formation of granular sludge led to the decrease of SV5 and SV30 in three SBRs (Fig. S2(a) and (b)). In the later stage of LP, the sedimentation of particles was slightly poor, because the growth of a large number of filamentous algae and ascomycotina reduced the particle density under long-term irradiation. The changes of biological species are described in detail later.
In order to quantify the amount of algae biomass in the granules, the content of Chl-a, Chl-b, and Chl-c were monitored. The main components of chlorophyll are Chl-a, and a small amount of Chl-c in three SBR reactors, which almost do not contain Chl-b (Fig. 1(d)). Because Chl-a is mainly found in green algae (Chlorophyta) and Chl-c is mainly found in diatom, dinoflagellate, brown algae (Phaeophyta), and Cryptophyta [30, 31]. The dominant species is Chlorophyta, the number of green algae is far greater than diatom, dinoflagellate, Phaeophyta, and Cryptophyta. This is consistent with the microscopic examination results introduced. Fig. 1(d) shows that Chl-c increases with the increase of illumination time at SP, and decreases at LP after 5 months of illumination. The decline may be due to the growth of other organisms as a result of light, creating inter-species competition that inhibits the growth of green algae. The concentration of dissolved oxygen (DO) in the central region of granules in dark, SP, and LP were 0.0003, 1.11, and 0.40 mg/L, respectively (Fig. 1(e)). There was no anaerobic zone (DO < 0.2 mg/L) in the algae-bacteria granules. For algae-bacteria granules containing algae, its internal DO content was significantly greater than aerobic granules. This property may be the reason leading to the change of microbial population. The living conditions of nitrite oxidizing bacteria and denitrifying bacteria are changed, and the number of populations decreases.
3.2. COD and P RemovalsCOD removal was better than 88% for all of the three SBRs during the whole reactors process with COD < 60 mg/L in effluent (Fig. S3). From week 11, the average removal rate of COD reached more than 90% till the end of the experiment. This phenomenon states clearly that the algal-bacterial granular system has a high capability of removing organics. Zhao et al. [32] also reported a similar phenomenon, about 90.6% of COD could be removed by mature algal-bacterial aerobic granular sludge. Different illumination times had no significant effect on COD removal by the algal-bacterial granules in the three SBRs.
The phosphate removal rates of all three SBRs approached 100% in the first seven weeks, but declined in the following five weeks, then increased and stabilized, fluctuating between 80 and 90% (Fig. S3). Generally, the fluctuation of phosphate removal in an SBRs is ascribed to the development of poly-phosphate accumulating organisms (PAOs) [33]. Bassin also reported that the phosphate release affected by PAOs, which dropped from 50 to 30 mgP/L, and the phosphate removal, which decreased from 80 to 30% [34]. The sedimentation at the later period is good, so sludge discharge was fluctuated, which may be the reason for the fluctuation of phosphorus removal rate.
3.3. N RemovalsThe variations of N species including NO2-N and NO3-N in the effluent, TN and NH4-N removal efficiency are illustrated in Fig. 2(b). The removal rates of ammonia nitrogen were above 96.4% in all three reactors (Fig. 2(b)). It is proved that both aerobic granular sludge and algae-bacteria granule have perfect treatment effect on ammonia nitrogen. The removal rate of TN increased from about 45% at the beginning to about 79% at the end. There was no significant difference among the three groups. In the three SBRs, NO3-N was the dominant N form, and only an extremely small amount of NO2-N (averagely 0.02 mg-N/L) was detected in the effluent. Dong et al. [35] detected that the nitrogen form in the effluent was mainly NO3-N, and only a very small amount of NO2-N with an average of 0.13 mg-N/L was detected under no salinity intrusion. NO2-N did not accumulate in the three SBRs from beginning to end, however, the NO3-N in the effluent of SBRs (dark, SP, and LP) from the initial 25 mg/L or so decrease to 12.78, 11.52, and 14.45 mg/L, respectively. The results of the above phenomena reflect that the three SBRs systems demonstrated NH4-N can be easily and effectively converted into NO3-N, and the nitrification effect is maintained.
Fig. 2 demonstrate that better nitrification and denitrification were always noticed in SP than in dark and LP during the whole operation, possibly due to its relatively suitable illumination time. Nevertheless, around 80% of the TN removal was through nitrification and denitrification, with the rest owing to biomass uptake [36]. This observation agrees with Kube et al. [37] that algae actively absorb or assimilate N and P, and transform them into their own nutrients for their own growth and reproduction, resulting in the increase of algae biomass [38]. Moreover, the rapid growth of algae leads to the increase of the pH value of the effluent (Fig. S4), which changes the physical and chemical properties of the water, promotes ammonia nitrogen volatilization and phosphate precipitation, so as to achieve the effect of nitrogen and phosphorus removal.
3.4. Lipid Content and Composition in the Granules Obtained at Different Illumination TimeThe lipid content and productivity are analyzed to discern the influence of illumination time on biosynthesis of lipid (Fig. 3), which is of great significance for the cultivation of algae-bacterial granules and the recovery of high value-added raw feedstock. The lipid content of the algae-bacteria granules in SP and LP was 19.62 and 33.71% higher than that of the bacterial granules (dark), respectively. The lipid content of algae is usually more than 10%, while the lipid content of sludge is about 6.4%. Generally, the lipid content of sludge is about 6.4% [39], so algae-bacteria have a greater contribution to the lipid content of the algae-bacteria granule.
There are 31 kinds of fatty acids in the bacterial granules form dark (Table S1). Among them, methyl 13-methylmyristate, methyl palmitate, and methyl trionylate accounted for 66.95% of the total fatty acids. In SP and LP, methyl pentadecanoate, methyl palmitate, methyl trionylate, and methyl 9-octadecenoic acid were the main species, accounting for 80.15% and 80.75%, respectively (Table S1, S2, and S3). It is noteworthy that LP did not drive significant changes in lipid composition, so SP was more suitable for algal bacteria symbiotic granules to produce lipid. According to the results of GC-MS analysis, saturated fatty acids account for 67, 51.6, and 53.3% of the particles of dark, SP and LP, respectively, with a larger proportion in dark (Fig. 3(b)). The monounsaturated fatty acids in SP and LP were about 43% higher than those in dark. Fig. 3(b) presents that there is little difference in the content of polyunsaturated fatty acids in the three SBRs. Obviously, strong illumination time favored the accumulation of mono-unsaturated fatty acid methyl esters (FAMEs) (C18:1) and poly-unsaturated FAMEs (C18:2) in the algal-bacterial granules, while negatively influenced the accumulation of saturated FAMEs. It has been reported that an increase in light intensity results in a significant increase in mono-unsaturated fatty acid [19, 40]. As we can see, the lipids produced consisted of over 61.35–61.15% (SP and LP) of saturated and mono-unsaturated FAMEs (palmitic acid esters (C16:0) and oleic acid esters (C18:1)), which are preferred components for biodiesel [41].
To further evaluate the most important properties of biodiesel derived from the algal lipid, kinematic viscosity (KV), iodine value (IV), cetane number (CN), and cold filter plugging point (CFPP) were estimated based on predictive models [42]. Table S2 and S3 indicate that the four biodiesel properties all satisfied the quality standards of Europe (EN), Germany, China, and the American Society for Testing and Materials (ASTM). However, as shown in Table S3, biodiesel prepared from algal bacterial granules (SP and LP) have higher quality and stability.
3.5. Analysis of Prokaryotic CommunityAs shown in Fig. 4, important difference in the proportion of dominant bacteria at the level of family and genus has been found in the granules of the three SBRs. The family of Rhodocyclaceae, Comamonadaceae, Thiotrichaceae, Propionibacteriaceae, and Saprospiraceae is about 30.5, 12.8, 3.4, 1.4, and 1.5%, occupying the largest portion of the bacteria in LP. While Rhodocyclaceae is only dominant family about 41.7% and 39.8% in dark and SP, respectively. The abundances of Rhodocyclaceae is the major microbial families involved in P removal [43–45]. The concentration of PO4-P in the effluent verified the P removal ability of dark and SP is slightly better than LP. Obviously, Fig. 4 demonstrates that light or algae growth will reduce the number of nitrosating bacteria (Nitrosomonadaceae). Nitrosomonadaceae was the predominant in partial nitrification algal-bacterial granule system, which was sensitive and unfavorable in the established system [46].
Zoogloea spp. dominating the granules in the three SBRs, relative abundance were about 38.4, 35.5, and 27.5%, respectively in dark, SP, and LP (Fig. 4(b)). Previously, it was found that Zoogloea spp. and Flavobacterium (0.13, 0.12, and 0.15% in dark, SP, and LP, respectively) were closely related to the formation of activated sludge flocculation and denitrification (Table S4) [47, 48]. Micrococcus spp. (micropruina, 0.2, 0.4, and 0.09% in dark, SP, and LP, respectively) has the ability to remove COD and ammonia nitrogen (Table S4) [49]. It should be noted that the proportion of Nitromonas in dark was 4.37%, which was much higher than that of SP and LP (2.75% and 0.75%) (Table S4). The content of Arthrobacter in dark and SP was 0.004%, which was not detected in LP (Table S4). The same trend also appears in the genus Zoocolloidal bacteria with denitrification function. Zoogloea spp. was reported to closely associate with the formation of activated sludge flocs and denitrification [47]. The content of Arthrobacter was very low in Dark and SP, but not detected in LP. The same trend also appears in the denitrification function of Zoogloea spp. The falling of above genus signaled that nitrifying and denitrifying bacteria were significantly inhibited under the symbiotic growth of bacteria and algae in SP and LP, resulting in decreased nitrification performance. This observation is to some extent in agreement with previous findings that the bacterial sludge or algae inhibit nitrification performance [11, 50]. However, in this study, the nitrification rate and denitrification rate were SP > LP > dark, indicating that algae played a key role in denitrification. Moreover, previous studies demonstrated that bacteria related to the Aquimonas, Azoarcur, and Thiothrix were shown to be abundant potential denitrifiers, which have been found in the granules of the three SBRs [51, 52]. In a word, the change in the biological community is not a simple physicochemical relationship, but a complex ecological relationship. Therefore, we can only analyze the bacterium as we known that are working about the removal of N, P, and COD. The changes in the community may be related to the intensity of light [53] and the allelopathy of the algae secretions [54]. It may also be related to algae changing the dissolved oxygen state of granules.
3.6. Analysis of Eukaryotic (including algae) communityIn aerobic granular sludge (dark), the rotifers were dominated in eukaryotes, up to 83.4%. However, the rotifers only contained 18.3 and 8.4% of the algae-bacteria granules in SP and LP, respectively (Fig. 5(a)). The reason is that light can promote the growth of algal, increase the abundance of eukaryotes, and cause species competition. However, for dark treatment, the competitiveness of species is very small, and rotifers are easy to break out. Diatomeas (Light-dependent organism) are able to apply the photoprotective mechanism to maintain high growth rates and a high photosynthetic efficiency over a wide range of light intensities [55–57]. Therefore, compared with SP, the relative abundance of Diatomea increased by 192.31% in LP, and long-term illumination promoted the rapid growth of Diatomea. Interestingly, In SP, the relative number of Chlorophyta was the largest, and high intensity light (LP) inhibited the growth of Chlorophyta [58]. Ascomycota accounted for 8.7% in LP and almost did not exist in the other two reactors. We have reason to consider that the source of many algae is due to a small amount of algae in the seed sludge, and also in tap water and air. In conclusion, suitable light can promote eukaryote abundance and facilitate lipid formation.
The difference in the level of the genus is basically the same as the level of the phylum (Fig. 5(b)). Rotifera include two genera (Adinetida and Rotaria). The main genus of Ascomycota was Knufia. The 18sDNA method was not able to identify a phylum (unidentified eukaryotes), which is Telotrochidium. The dominant genus of chlorophyta was Caespitella, which have been renamed to Stigeoclonium. The dominant genus of Diatomea was Navicula. There are also some dominant genera of Ciliatea which belong to protozoa, such as Trochilia and Hemiophrys. The 18sDNA method is not able to identify algae to the level of the genus or species. The results of microscopic identification of algae are shown in Table 1. Navicula cuspidate, Navicula oblonga, Melosira varians, Aulacodiscus sp., and Navicula cryptocephala belong to the phylum of Diatomea. Stigeoclonium fasciculare, Chlorella vulgaris, Chlorella ellipsoidea, Actinastrum sp., and Hyalotheca dissiliens belong to the phylum of Chlorophyta. Planktolyngbya subtilis, Anabeana cylindrical, and Merismopedia sp. belong to the phylum of Cyanobacteria. Glenodinium pulvisculus belong to the phylum of Dinophyta. In addition, according to microscopic examination, Navicula spp. and Chlorella spp. are distributed in the interior of granule, while filamentous green algae and bacterium are distributed in the outer layer of granule. Arcila et al. [13] had similar finding in anaerobic treatment systems.
4. ConclusionsThe formation of algae-bacteria granule is an effective way to reduce harvest cost. Microbial community can be significantly influenced by different photoperiod which was perceived as a selective pressure. In short photoperiod (12 h/d) sludge can generate symbiotic algae-bacteria granules in the SBR. Instead, long photoperiod (24 h/d) promotes the growth of algae and Ascomycota early, and then inhibits algae, especially Chlorophyta. In the end, algae-bacteria-fungi granules were formed.
By using GC-MS and gravimetric method, it was found that the lipid content of algae-bacteria granules is 33.71% more than that of pure aerobic granular sludge. However, the proportion of unsaturated fatty acids (46.67–48.38%) in algae-bacteria granules is higher, resulting in the quality of biodiesel prepared from algae-bacteria granules is slightly worse than that prepared from pure aerobic granular sludge. But both of them were better than that biodiesel prepared from pure algae. This study provides new insights into the strategy of enriching lipid in algal bacterial culture and supplements the information on microbial ecology and diversity in algal-bacterial culture. Moreover, nitrospiraceae was sensitive and unfavorable in the established system. The mechanism of interaction between bacteria and algae remains to be further studied.
AcknowledgmentsThis study was financially supported by the Fundamental Research Funds for the Central Universities and the China Postdoctoral Science Foundation (2020M680868) and National Natural Science Foundation of China (grants 51608279).
NotesAuthor Contributions X.L.M. (Intermediate engineer) and G.S.G. (Research assistant) conducted all the experiments and wrote the manuscript. H.W.L. (Professor) and S.B.B. (Research assistant) revised the manuscript. M.F.S. (Instructor) and G.S.G. (Research assistant) wrote and revised the manuscript. References1. Lananan F, Abdul Hamid SH, Din WNS, et al. Symbiotic bioremediation of aquaculture wastewater in reducing ammonia and phosphorus utilizing Effective Microorganism (EM-1) and microalgae (Chlorella sp.). Int Biodeter Biodegr. 2014;95:127–134.
2. Su Y, Mennerich A, Urban B. Municipal wastewater treatment and biomass accumulation with a wastewater-born and settleable algal-bacterial culture. Water Res. 2011;45:3351–3358.
3. Posadas E, Garcia-Encina PA, Soltau A, Domínguez A. Carbon and nutrient removal from centrates and domestic wastewater using algal-bacterial biofilm bioreactors. Bioresour Technol. 2013;139:50–58.
4. Isanta E, Suárez-Ojeda ME, ValdelRío Á, Morales N, Perez J, Carrera J. Long term operation of a granular sequencing batch reactor at pilot scale treating a low-strength wastewater. Chem Eng J. 2012;198–199:163–170.
5. Yan L, Zhang M, Liu Y, et al. Enhanced nitrogen removal in an aerobic granular sequencing batch reactor under low DO concentration: Role of extracellular polymeric substances and microbial community structure. Bioresour Technol. 2019;289:121651.
6. Ramanan R, Kim BH, Cho DH, Oh HM, Kim HS. Algae-bacteria interactions: Evolution, ecology and emerging applications. Biotechnol Adv. 2016;34:4–29.
7. Hoh D, Watson S, Kan E. Algal biofilm reactors for integrated wastewater treatment and biofuel production: A review. Chem Eng J. 2016;287:466–473.
8. Liu L, Hong Y, Ye X, et al. Biodiesel production from microbial granules in sequencing batch reactor. Bioresour Technol. 2018;249:908–915.
9. Liu L, Zeng Z, Bee M, et al. Characteristics and performance of aerobic algae-bacteria granular consortia in a photo-sequencing batch reactor. J Hazard Mater. 2018;349:135–142.
10. Meng F, Liu D, Huang W, Lei Z, Zhang Z. Effect of salinity on granulation, performance and lipid accumulation of algal-bacterial granular sludge. Bioresour Technol Reports. 2019;7:100228.
11. Huang W, Li B, Zhang C, et al. Effect of algae growth on aerobic granulation and nutrients removal from synthetic wastewater by using sequencing batch reactors. Bioresour Technol. 2015;179:187–192.
12. Li B, Huang W, Zhang C, et al. Effect of TiO2 nanoparticles on aerobic granulation of algal-bacterial symbiosis system and nutrients removal from synthetic wastewater. Bioresour Technol. 2015;187:214–220.
13. Arcila JS, Buitrón G. Influence of solar irradiance levels on the formation of microalgae-bacteria aggregates for municipal wastewater treatment. Algal Res. 2017;27:190–197.
14. Ajabshir SZ, Kamazani MM. Recent advances in nanostructured Sn–Ln mixed-metal oxides as sunlight-activated nanophotocatalyst for high-efficient removal of environmental pollutants. Ceram Int. 2021;47:23702–23724.
15. Ajabshir SZ, Asil SA, Niasari MS. Simple and eco-friendly synthesis of recoverable zinc cobalt oxide-based ceramic nanostructure as high-performance photocatalyst for enhanced photocatalytic removal of organic contamination under solar light. Sep Purif Technol. 2021;267:118667.
16. Safajou H, Ghanbari M, Amiri O, et al. Green synthesis and characterization of RGO/Cu nanocomposites as photocatalytic degradation of organic pollutants in waste-water. Int J Hydrog Energy. 2021;46:20534–20546.
17. Ajabshir SZ, Baladi M, Niasari MS. Enhanced visible-light-driven photocatalytic performance for degradation of organic contaminants using PbWO4 nanostructure fabricated by a new, simple and green sonochemical approach. Ultrason Sonochem. 2021;72:105420.
18. Ajabshir SZ, Morassaei MS, Amiri O, Salavati-Niasari M. Green synthesis of dysprosium stannate nanoparticles using Ficus carica extract as photocatalyst for the degradation of organic pollutants under visible irradiation. Ceram Int. 2020;46:6095–6107.
19. Meng F, Xi L, Liu D, et al. Effects of light intensity on oxygen distribution, lipid production and biological community of algal-bacterial granules in photo-sequencing batch reactors. Bioresour Technol. 2019;272:473–481.
20. Huang W, Cai W, Huang H, et al. Identification of inorganic and organic species of phosphorus and its bio-availability in nitrifying aerobic granular sludge. Water Res. 2015;68:423–431.
21. Adav SS, Lee DJ, Tay JH. Extracellular polymeric substances and structural stability of aerobic granule. Water Res. 2008;42:1644–1650.
22. Piasecka A, Krzemińska I, Tys J. Physical methods of microalgal biomass pretreatment. Int Agrophys. 2014;28:341–348.
23. Hu HJ, Wei YX. The Freshwater Algae of China: Systematics. Taxonomy and Ecology. Pekin: Science Press; 2006.
24. Ho TW, Hwang JS, Cheung MK, Kwan HS, Wong CK. DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J Exp Mar Biol Ecol. 2017;494:1–9.
25. Elwood HJ, Olsen GJ, Sogin ML. The small-subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol Biol Evol. 1985;2:399–410.
26. Ritchie RJ. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica. 2008;46:115–126.
27. Amin SA, Hmelo LR, Vantol HM, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101.
28. Milferstedt K, Kuo-Dahab WC, Butler CS, et al. The importance of filamentous cyanobacteria in the development of oxygenic photogranules. Sci Rep. 2017;7:1–15.
29. Zhang B, Guo Y, Lens PNL, et al. Effect of light intensity on the characteristics of algal-bacterial granular sludge and the role of N-acyl-homoserine lactone in the granulation. Sci Total Environ. 2019;659:372–383.
30. García F, Freile-Pelegrín Y, Robledo D. Physiological characterization of Dunaliella sp. (Chlorophyta, Volvocales) from Yucatan, Mexico. Bioresour Technol. 2007;98:1359–1365.
31. Jerez CG, Malapascua JR, Sergejevová M, Masojídek J, Figueroa FL. Chlorella fusca (Chlorophyta) grown in thin-layer cascades: Estimation of biomass productivity by in-vivo chlorophyll a fluorescence monitoring. Algal Res. 2016;17:21–30.
32. Zhao Z, Yang X, Cai W, et al. Response of algal-bacterial granular system to low carbon wastewater: Focus on granular stability, nutrients removal and accumulation. Bioresour Technol. 2018;268:221–229.
33. Monclúsa H, Sipmaa J, Ferrero G, Rodriguez-Roda I, Comas J. Biological nutrient removal in an MBR treating municipal wastewater with special focus on biological phosphorus removal. Bioresour Technol. 2010;11(101)3984–3991.
34. Bassin JP, Kleerebezem R, Dezotti M, van Loosdrecht MCM. Simultaneous nitrogen and phosphate removal in aerobic granular sludge reactors operated at different temperatures. Water Res. 2012;46:3805–3816.
35. Dong X, Zhao Z, Yang X, et al. Response and recovery of mature algal-bacterial aerobic granular sludge to sudden salinity disturbance in influent wastewater: Granule characteristics and nutrients removal/accumulation. Bioresour Technol. 2021;321:124492.
36. Xu M, Li P, Tang T, Hu Z. Roles of SRT and HRT of an algal membrane bioreactor system with a tanks-in-series configuration for secondary wastewater effluent polishing. Ecol Eng. 2015;85:257–264.
37. Kube M, Jefferson B, Fan L, Roddick F. The impact of wastewater characteristics, algal species selection and immobilisation on simultaneous nitrogen and phosphorus removal. Algal Res. 2018;31:478–488.
38. Ogbonna JC, Yoshizawa H, Tanaka H. Treatment of high strength organic wastewater by a mixed culture of photosynthetic microorganisms. J Appl Phycol. 2000;12:277–284.
39. Zhu F, Zhao L, Jiang H, et al. Comparison of the lipid content and biodiesel production from municipal sludge using three extraction methods. Energ Fuel. 2014;28:5277–5283.
40. Krzemińska I, Piasecka A, Nosalewicz A, Simionato D, Wawrzykowski J. Alterations of the lipid content and fatty acid profile of Chlorella protothecoides under different light intensities. Bioresour Technol. 2015;196:72–77.
41. Sheehan J, Dunahay T, Benemann J, Roessler P. A look back at the US Department of Energy’s aquatic species program: biodiesel from algae. United States: National Renewable Energy Lagoratory; 1998. p. 1–294.
42. Song M, Pei H, Hu W, Ma G. Evaluation of the potential of 10 microalgal strains for biodiesel production. Bioresour Technol. 2013;141:245–251.
43. Yae JB, Ryu JH, Tuyen VN, Kim HG, Hong SW, Ahn DH. An aerobic granular sludge process for treating low carbon/nitrogen ratio sewage. Environ Eng Res. 2019;24:238–245.
44. Kong Y, Nielsen JL, Nielsen PH. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants. Appl Environ Microb. 2005;71:4076–4085.
45. Ge H, Batstone DJ, Keller J. Biological phosphorus removal from abattoir wastewater at very short sludge ages mediated by novel PAO clade Comamonadaceae. Water Res. 2015;69:173–182.
46. Huang W, Liu D, Huang W, Cai W, Zhang Z, Lei Z. Achieving partial nitrification and high lipid production in an algal-bacterial granule system when treating low COD/NH4–N wastewater. Chemosphere. 2020;248:126106.
47. Shao Y, Chung BS, Lee SS. Zoogloea caeni sp. nov., a floc-forming bacterium isolated from activated sludge. Int J Syst Evol Microbiol. 2009;59:526–530.
48. Xiao J, Guo P, Huo W, Yu J, Zhu C. Application of denitrifying microbes to wastewater denitrification. Environ Sci Technol (China). 2009;32(12)97–102.
49. Li J, Ji S, Liu Z, Qin Z, Liu Y, Yang Y. Analysis of bacterial composition of aerobic granular sludge with 16S rDNA clone library. Res Environ Sci. 2009;22:1218–1223.
50. Liu L, Fan H, Liu Y. Development of algae-bacteria granular consortia in photo-sequencing batch reactor. Bioresour Technol. 2017;232:64–71.
51. Thomsen TR, Kong Y, Nielsen PH. Ecophysiology of abundant denitrifying bacteria in activated sludge. FEMS Microbiol Ecol. 2007;60:370–382.
52. Thomsen TR, Nielsen JL, Ramsing NB, Nielsen PH. Micromanipulation and further identification of FISH-labelled microcolonies of a dominant denitrifying bacterium in activated sludge. Environ Microbiol. 2004;6:470–479.
53. Wang M, Yang H, Ergas SJ, et al. A novel shortcut nitrogen removal process using an algal-bacterial consortium in a photo-sequencing batch reactor (PSBR). Water Res. 2015;87:38–48.
54. Zhou D, Li Y, Yang Y, Wang Y, Zhang C, Wang D. Granulation, control of bacterial contamination, and enhanced lipid accumulation by driving nutrient starvation in coupled wastewater treatment and Chlorella regularis cultivation. Appl Microbiol Biotechnol. 2015;99:1531–1541.
56. Lavaud J, Rousseau B, Etienne AL. Enrichment of the light-harvesting complex in diadinoxanthin and implications for the nonphotochemical fluorescence quenching in diatoms. Biochem. 2003;42:5802–5808.
Fig. 1Morphology of granular sludge (a), variations of MLVSS (b), SVI (c), chlorophy (d), and DO (e) concentrations during the operation of the three SBRs. 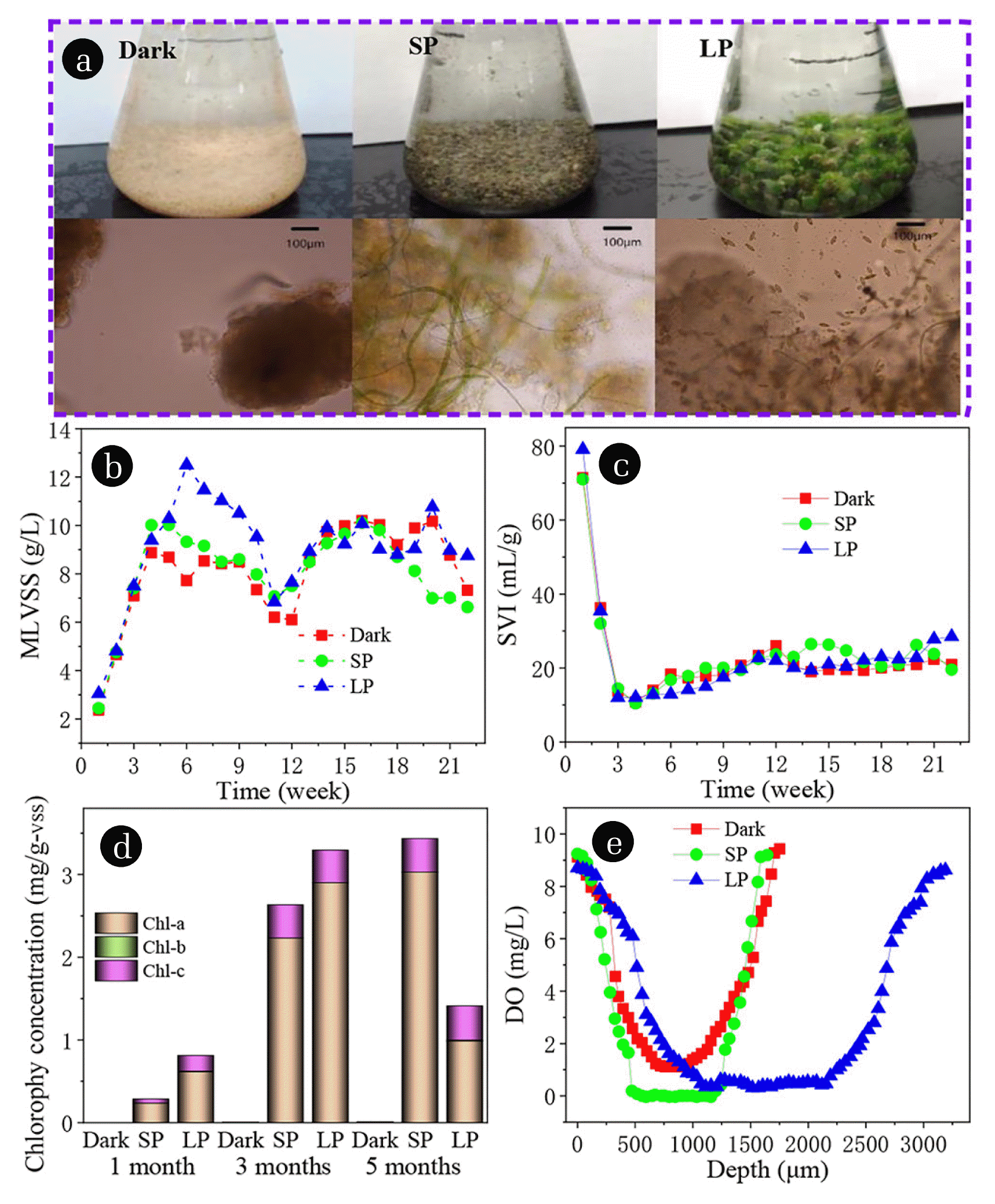
Fig. 2Variations of nitrification and denitrification rate (a), NO4+-N & TN removal rate and NO3-N & NO2-N concentrations (b) during the operation of the three SBRs. 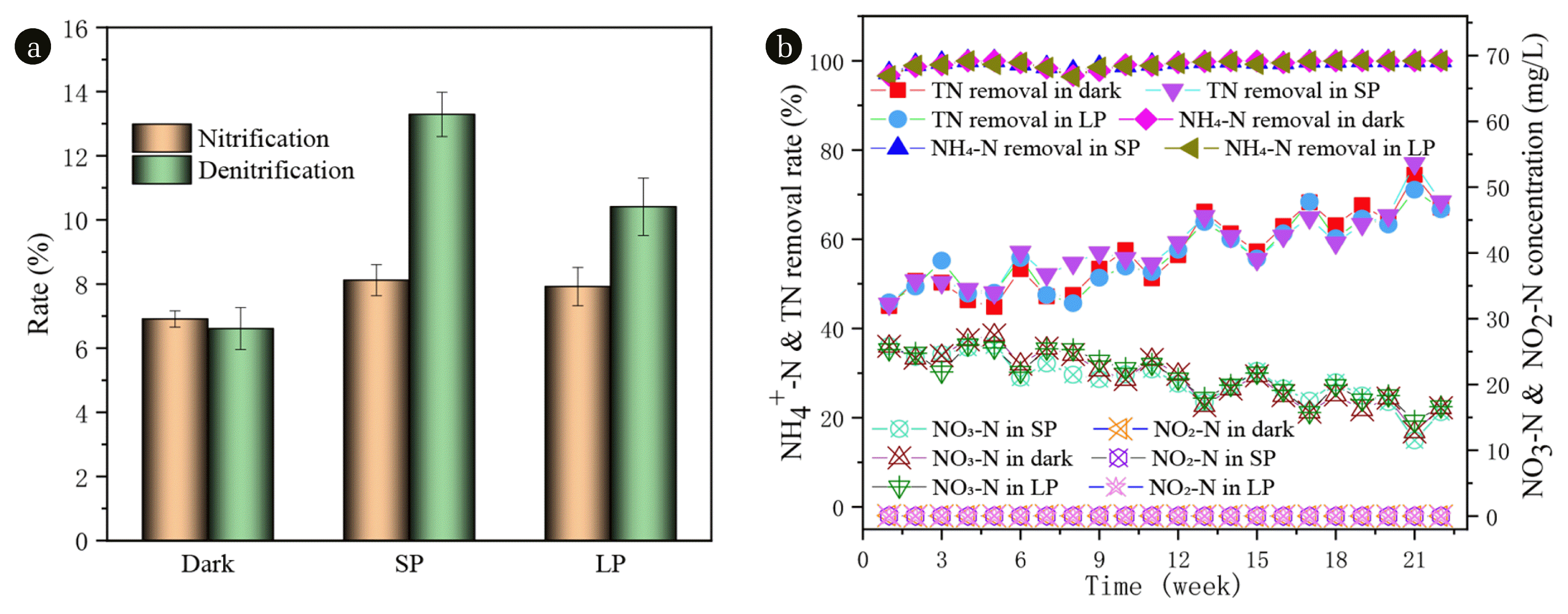
Table 1The Species of Algae in Three Reactors and Its Relative Abundance |
|
|||||||||||||||||||||||||||||||||||||