1. Introduction
Carbofuran (2,3-dihydro-2,2-dimethyl-7-benzofuranyl N-methylcarbamate) is the most toxic class I carbamate widely used as an insecticide, nematicide, and acaricide [1]. Though easily hydrolyzed, this compound is fairly persistent in soils [1]. It exhibits high mammalian toxicity due to neural impairment through cholinesterase inhibition [2]. The compound is of high mobility in soil [3], and its residues have been detected in soils and water sources [4]. Besides, the pesticide contaminates fresh vegetables causing food contamination [5]. As a result, carbofuran has been banned from agricultural use in the US and EU, but it is still widely used in developing countries [6].
Hence, it is necessary to remove carbofuran from contaminated environments. Rather than using physical and chemical methods, microorganisms are more effective and environmentally friendly in treating this chemical. There are several microbial isolates which can degrade carbofuran such as Aspergillus niger and Fusarium graminearum [7], Arthrobacter, Pseudomonas, Bacillus, and Actinomyces [8], Pseudomonas sp. [9], Sphingomonas sp. SB5 [10], Novosphingobium sp. [11], Burkholderia cepacia [12], Cupriavidus sp. [13]. Also, the immobilization of microorganisms in rice plant components has been reported in some previous research [6, 14]. However, no study describing carbofuran degradation in liquid media by biofilm has been mentioned in the reported literature so far.
It is widely known that rice straw is a huge by-product available in agricultural countries around the world. This by-product material quickly makes a good place to live and lay eggs for insects. Thus, farmers often burn it to eliminate their infectious rapid spreading, causing severe air pollution. Rice straw is a highly promising natural and biodegradable material used in soil bioaugmentation for cell immobilization. It has an amendment for the improvement of physical properties and fertilization [15]. In addition, by applying effective microorganisms, the composting material has enhanced nutrient values [16]. Castro-Gutiérrez et al. [17] showed that biomixture of coconut fiber, garden compost and soil pre-exposed to carbofuran could be used to depurate pesticide. However, the use of rice straw for immobilizing bacteria and carbofuran biodegradation in soil has not been documented yet.
Thus, the present research aims to (1) isolating and identifying carbofuran-degrading bacteria, (2) investigating the effects of co-substrates on biofilm formation and isolated strain biodegradation, (3) examining biodegradation of carbofuran in soil by bacteria immobilized in rice straw versus free cells, and (4) determining degradation metabolites for the compound.
2. Materials and Methods
2.1. Culture Media
The mineral medium (MM) with the components of 2.79 mg/L Na2HPO4, 1.0 mg/L KH2PO4, 0.2 mg/L MgSO4·H2O, and 1.0 mL of trace mineral solution was used for bacterial isolation, chemical degradation, and biofilm formation. The trace mineral solution consisted of H3BO3, 0.30; CoCl2·6H2O, 0.20; ZnSO4·7H2O, 0.10; Na2MoO4·2H2O, 0.03; MnCl2·4H2O, 0.03; NiCl2·6H2O, 0.02; CuCl2·2H2O, 0.01 in grams per liter [18]. Glucose and ammonium sulfate with different concentrations, ranging from 0.05 to 0.2 mM were added to MM for performing biofilm experiments. The media were autoclaved at 121°C for 15 min. All the applied chemicals with premium purity grades were purchased from either Sigma-Aldrich (Singapore) or Merck (Germany). Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract and 10 g/L NaCl) was also used for bacteria cultivation.
2.2. Enrichment, Isolation, and Identification of Carbofuran-Degrading Bacteria
Soil samples were collected from several random rice field sites in Dong Thap Province, Viet Nam. The 500 g soil samples were then transferred to a plastic container (length × width × depth: 15 cm × 25 cm × 20 cm). Distilled water was added to the soil on the basis of 40% water holding capacity. The container was capped with a plastic cover and kept intact for one month during the enrichment. Carbofuran was supplemented with a concentration of 120 mg/L periodically in every 5-day. Consequently, 5.0 g soil was dispensed in a 500 mL flask containing 200 mL of MM and 120 mg/L carbofuran. The flask was incubated with a speed of 150 rpm for 1 month, and 120 mg/L carbofuran was supplemented periodically in every 5-day. All the experiments were carried out at normal room temperature (around 30 °C). The sample solution was diluted and spread on solid MM (2% agar), supplemented with 120 mg/L carbofuran as an exclusive source of carbon and energy. Then, single colonies were transferred to the liquid MM and added with 120 mg/L carbofuran to examine their growth and biodegradability. Genomic DNA samples were extracted using an InstaGenetm Matrix (BIO-RAD, Hercules, USA). The 16S rRNA gene fragment was PCR amplified using the universal primers: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) at 95°C for 2 min and then 35 cycles of 95, 55 and 72°C for 1 min each, followed by 10 min at 72°C. The purification of amplification products was carried out using a multiscreen filter plate (Millipore Corp., Bedford, MA, USA). The obtained gene sequences of 16S rRNA were compared with other sequences available in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and identified using Ribosomal Database Project (RDP; http://rdp.cme.msu.edu).
ClustalW algorithm was used to aligne 16s rRNA sequences. 16s rRNA sequences were aligned by ClustalW algorithm. Tajima-Nei model and bootstrap method with 1,000 replications in MEGA X software were used to build the phylogenetic tree.
2.3. Carbofuran Degradation by Freely Suspended Cells
Chemical degradation by freely suspended cells at differential concentrations of carbofuran (60, 100, 120 and 180 mg/L) was performed in the MM. Bacteria were first cultured for 24 h in a liquid medium, then 1.0 mL of them was transferred to 100 mL of respective media to give 106 cells/mL, which was used to determine carbofuran degradation and biofilm formation. Liquid samples were collected to determine the cell growth and carbofuran residues during the incubation process.
2.4. Biofilm Formation Assay
Biofilm formation and consequent degradation of substrate were conducted in the reactor system as described by Nguyen and Ha [19]. The system comprised 150-mL cylindrical bottles with seven pieces of microscope glass slides (dimension: 24.5 mm × 76.2 mm × 0.8 mm) embedded separately into plastic devices stably stuck to the cover. Then, 50 mL of MM supplemented with glucose and ammonium sulfate (0.1% each) was added to each bottle in which 3/4 of the glass slides were submerged in the liquid media under a static condition. Carbofuran was also added at 120 mg/L. The incubation processes were conducted at room temperature with a shaking speed of 60 rpm. The formation of biofilm on the glass slides was quantitated based on the feasible counts of attachment cells as described in a previous study [19].
2.5. Carbofuran Degradation in the Biofilm-Batch Reactors
The biofilm reactor and incubation conditions have been specified above. After 24 h of time, the medium was removed. All the bottles and slides were rinsed in triplicate with sterilized saline solution (0.85% NaCl), filled with a new medium with 120 mg/L carbofuran and incubation process continued for 24 h. Bacteria in biofilm were then used for biodegradation while without bacteria were run in parallel as controls for the comparison.
The degradation was also carried using bacteria immobilized in rice straw. Rice straw randomly collected from a rice field immediately after harvest was also used for cell immobilization. The rice straw was first washed with distilled water and then cut into pieces (2 cm in length). The fresh straw whose total outside area was equal to that of the glass slides (~ 78.41 cm2) was dried in an oven (Ecocell-LIS-B2V/EC55, Germany) at 60°C for 2 d, and sterilized for subsequent immobilization of bacteria. The biofilm formation on the straw was carried out similar to the process on glass slides as described above. The chemical components in rice straw were analyzed according to the protocols described in the APHA method [20].
2.6. Carbofuran Biodegradation in Soil
The soil sample was collected from a depth of 10–50 cm in Dong Thap University campus, whose overall land has never been contaminated with pesticides. The soil was then air-dried at room temperature, sieved through a mesh with a diameter of 2 mm. The soil components were determined as 47.8% sand, 22.5% silt and 29.7% clay according to the soil texture triangle [21]. Other physicochemical properties were analyzed using the APHA method [20].
For carbofuran degradation in soil, 500 g soil with 40% of the water holding capacity was transferred into a container as described above. The soil was spiked with carbofuran and closed with a plastic cover. The container was incubated for 4 months for the natural degradation, and carbofuran was supplemented in every 1.0-month period at 1.5 mg/kg soil. The carbofuran residue in the soil at the last cycle was found insignificant. This soil was considered as history insecticide-treated soil. Consequently, history treated and non-treated soils (collected from the same site) with the same chemical and physical properties were used for determining carbofuran degradation. These soils were supplemented with 10 mg/kg of carbofuran and incubated for 30 d. Distilled water was also added for making the moisture content of approximately 40%.
For carbofuran biodegradation in soil by free cells, bacteria were cultured in LB for 24 h, collected by centrifuging it with a speed of 8,000 rpm for 5 min, washed twice with saline water and re-suspended in distilled water with approximately 5 × 107 CFUs/mL. The cell suspension was added in the soil to give a final concentration of 106 cells/g dry soil for both free and immobilized cell treatments and 40% of soil moisture.
For chemical degradation by immobilized cells, 5.0 g dry straw was cut into 1–2 mm in size, put in a sterile beaker and added with 50 mL of cell suspension (A). The mixture was then mixed with a plastic device and held in a laminar airflow cabinet for 18 h so that most liquid medium outside the straw got dried and the bacteria were absorbed by the straw. The straw was then gently rinsed with 10.0 mL of saline water (0.85% NaCl) in the beaker to remove unimmobilized cells (B). The difference between the cell numbers in cell suspension (A) and unimmobilized cells (B) in saline determined the cell density immobilized in the supporting material. The immobilized cells were found to be approximately 107 CFU/g dry material. After that, the straw was thoroughly mixed with soil.
To extract carbofuran from the soil, 5.0 g soil sample with or without straw was collected and pulverized using a mortar and pestle. The sample was then transferred to a glass tube, added with 15 mL of methanol and shaken horizontally with a speed of 150 rpm for 20 min. This process was carried out twice. The extract was filtered with a 0.22 μm syringe filter, evaporated under a nitrogen stream. The residue was then dissolved in methanol. In this process, the recovery efficiency of carbofuran from the soil was 95.6%.
2.7. Determination of Microbial Adherence to Hydrocarbon (MATH)
The bacteria cultured in the respective media were collected by doing centrifuge at 8,000 rpm for 5 min. Cell pellets were washed twice with saline water and re-suspended in phosphate urea magnesium buffer with the components as described in a previous report [22] and adjusted to OD600 equal to 0.5. The hydrophobicity of bacterial surface was determined using the MATH test. 4.0 mL of cell suspension was mixed with 1.0 mL of n-hexadecane in a glass test tube. The mixture was vigorously rotated for 2 min and left standing for 20 min. The aqueous phase was removed using a micropipette. The cell suspension partition was assessed as the percentage of bacteria adsorbed by a liquid hydrocarbon, and the cell-free buffer was served as the blank. The adherence percentage was measured using the formula: (1 − A/A0) × 100, where A0 and A are the OD600 of the bacterial suspension before and after mixing, respectively.
2.8. Determination of Degradation Metabolites and Analytical Methods
For identification of degradation metabolites, chemicals were extracted from liquid media as described by Plangklang and Reungsang [12] with some modification. 5.0 mL of culture media was added with 5.0 mL of absolute methanol and was shaken with a speed of 150 rpm for 20 min. Carbofuran and its intermediates were then extracted with the same volume of dichloromethane. The organic fraction was collected and evaporated to dryness under a nitrogen stream before dissolving in methanol. These processes were carried out twice. The degradation products were determined using gas chromatography-mass spectrometry (GC-MS, Agilent 7890N/MS 5975), while the residues during the degradation process were measured using HPLC as described in a previous publication [19]. Besides, cell turbidity and exponential growth rate in liquid media were determined according to Duc [18].
2.9. Statistical Analysis
All experiments were carried out in triplicate. The data obtained from the present studies were shown as the means ± standard deviation. The SPSS software program version 22.0 was used to analyze variance and calculate the significant differences (p < 0.05) using Duncan’s test.
3. Results and Discussion
3.1. Isolation and Identification of the Carbofuran-Degrading Strain
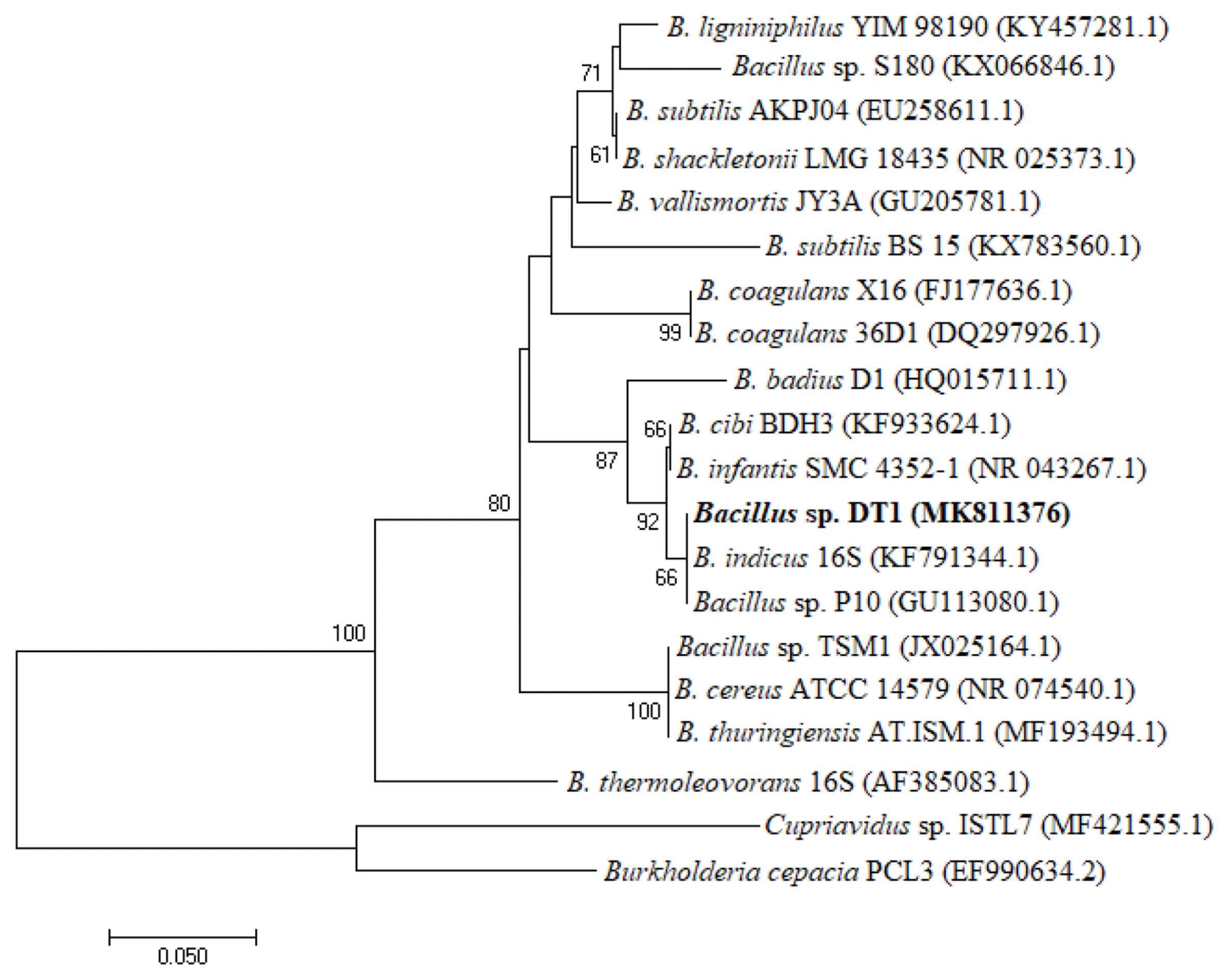
After enrichment and isolation, several carbofuran-degrading bacterial strains named DT1, DT2, DT3, and DT4 were obtained. Only strain DT1 could utilize carbofuran as a sole carbon and energy source, and no isolate utilized the compound as a nitrogen source. Other strains degraded the herbicide if the media were added with a carbon source such as glucose and succinate and a nitrogen source such as sodium nitrate and ammonium sulfate (data not shown). Therefore, the strain DT1 was used in other experiments. While comparing 16S rRNA obtained from other sequences available in the NCBI GenBank, it shows that the strain identified a close homology with other Bacillus strains such as Bacillus sp. P10 (99.44%), Bacillus indicus 16S (99.30%), and Bacillus cibi BDH3 (99.30%). The RDP analysis also places this strain in genus Bacillus. Hence, the isolated substance was named as Bacillus sp. DT1. The phylogenetic tree was constructed based on 16S rRNA shown in Fig. 1. The DT1 16S rRNA was deposited in the GenBank with accession N0. MH109504.1. The bacterial sample was also stored in the Culture Collection at the Center for Biochemical Analysis (Vietnam) with deposition N0. DUCOANH2015-6DT1.
3.2. Biodegradation of Carbofuran in Liquid Culture by Freely Suspended Bacillus sp. DT1
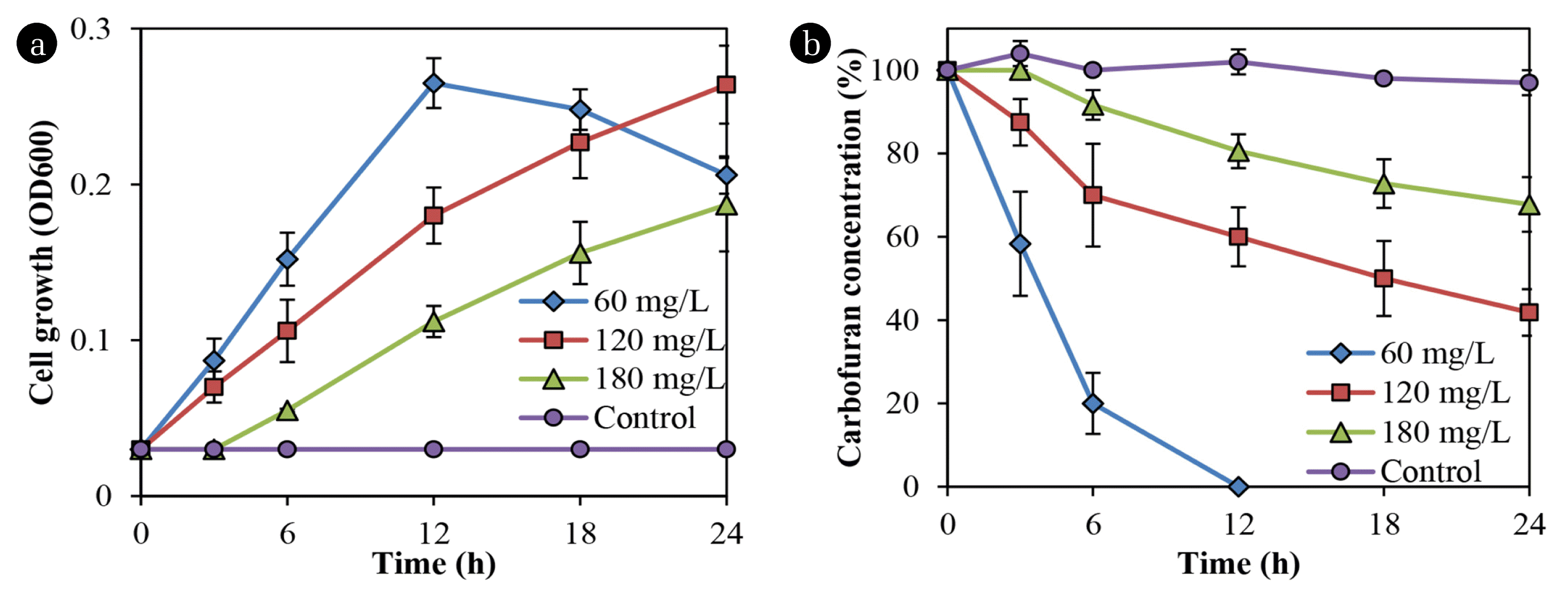
Fig. 2 shows that Bacillus sp. DT1 utilized carbofuran as a sole carbon and energy source at varying rates. The isolated strain completely utilized the substrate at the concentration of 60 mg/L within 12 h. However, an increase in carbofuran concentrations reduced in degradation and cell growth rates. Meanwhile, no degradation was found in the abiotic controls.
In previous studies, the degradation in liquid and soil media by pure cultures have been conducted, but only some microbial isolates showed effective degradation of the pesticide [13]. For example, Gupta et al. [13] reported that Cupriavidus sp. ISTL7 could effectively degrade the compound with approximately 98% of 400 ppm within 96 h. The effects of carbofuran on the production of extracellular polymeric substances by Cupriavidus sp. ISTL7 was also reported, but the biofilm formation and degradation in soil were not determined [13].
3.3. Effects of Co-substrates on Biofilm Formation and Carbofuran Degradation
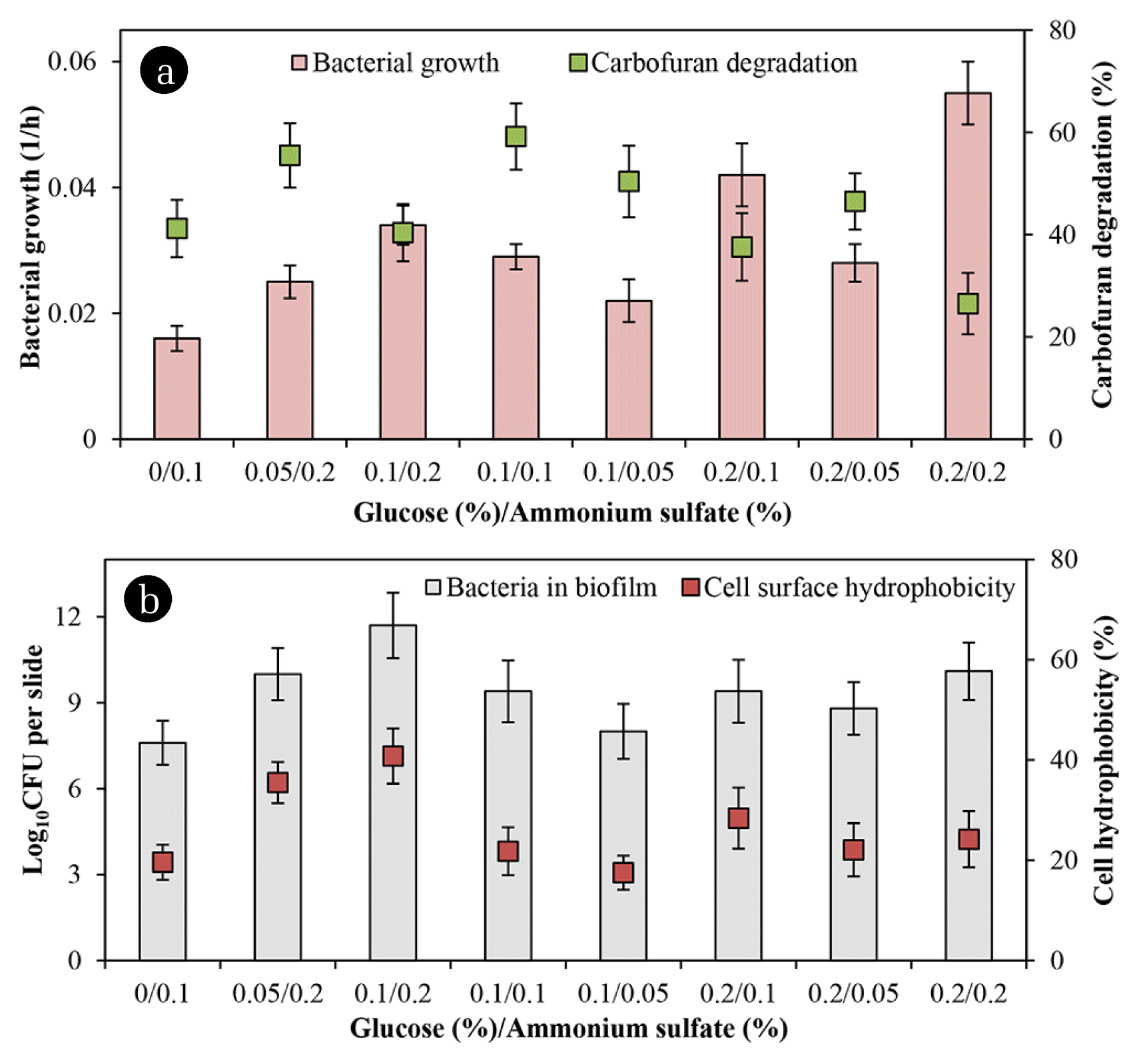
Fig. 3(a) presents that the rate of degradation increased when the glucose/ammonium sulfate ratio of 0.05/0.2 and 0.1/0.1 was added. It also shows that any supplemented co-substrates stimulated cell numbers in both biofilm and the suspended counterpart. However, in presence of glucose/ammonium sulfate ratio of 0.2/0.1 and 0.2/0.2 resulted in higher growth rates, but lower the chemical degradation because the bacteria appeared to utilize these nutrients rather than carbofuran. Besides, more ammonium sulfate in the media seemed to boost biofilm development while the numbers of attached cells and cell hydrophobicity were closely proportional as presented in Fig. 3(b). The bacterial attachment on supporting material closely related to its surface hydrophobicity. However, the degradation did not relate to cell hydrophobicity in the present study. Related literature has already documented that cell surface hydrophobicity and nutrients positively influence biofilm development [22, 23]. Besides, cell surface hydrophobicity is a key component in the biodegradation of organic compounds and biofilm formation [22]. Also, cell surface hydrophobicity is related to microbial degradation of 4-nitrotoluene [24] and pyrene [25].
3.4. Degradation of Carbofuran by Bacteria Immobilized on Glass Slides and Rice Straw
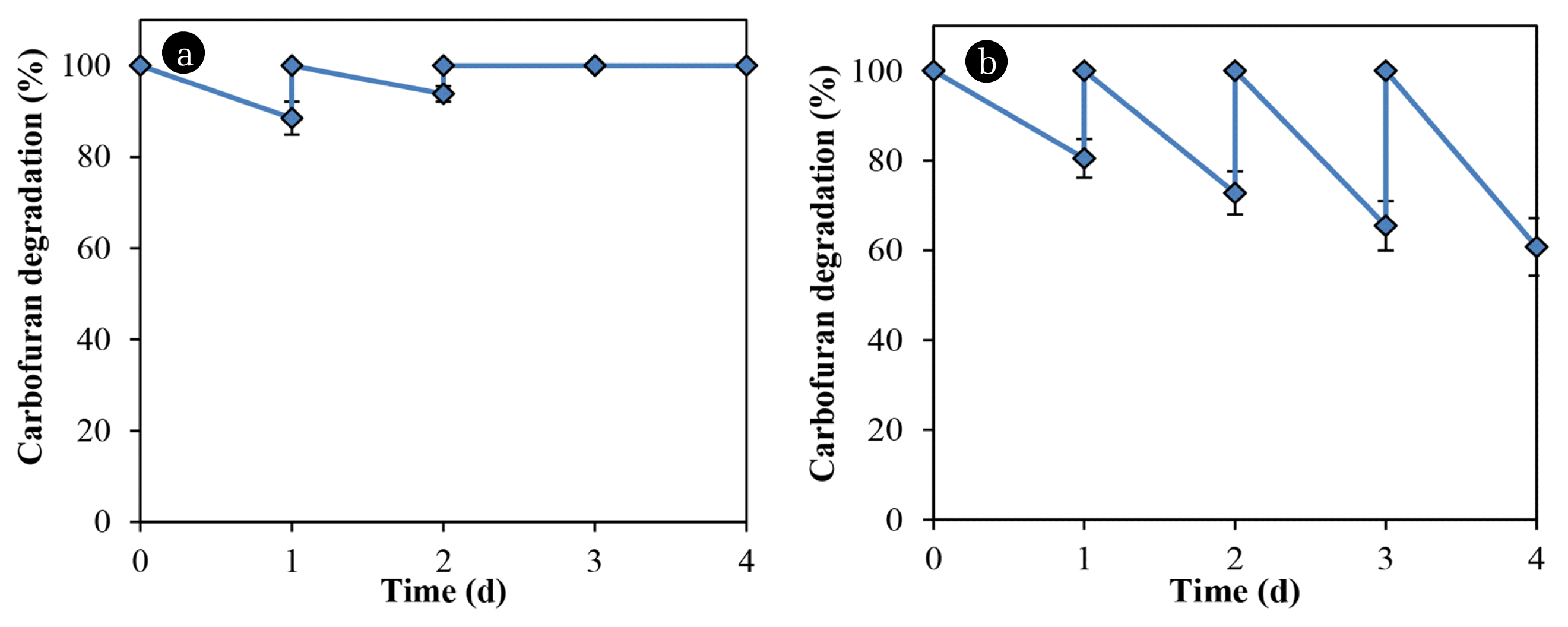
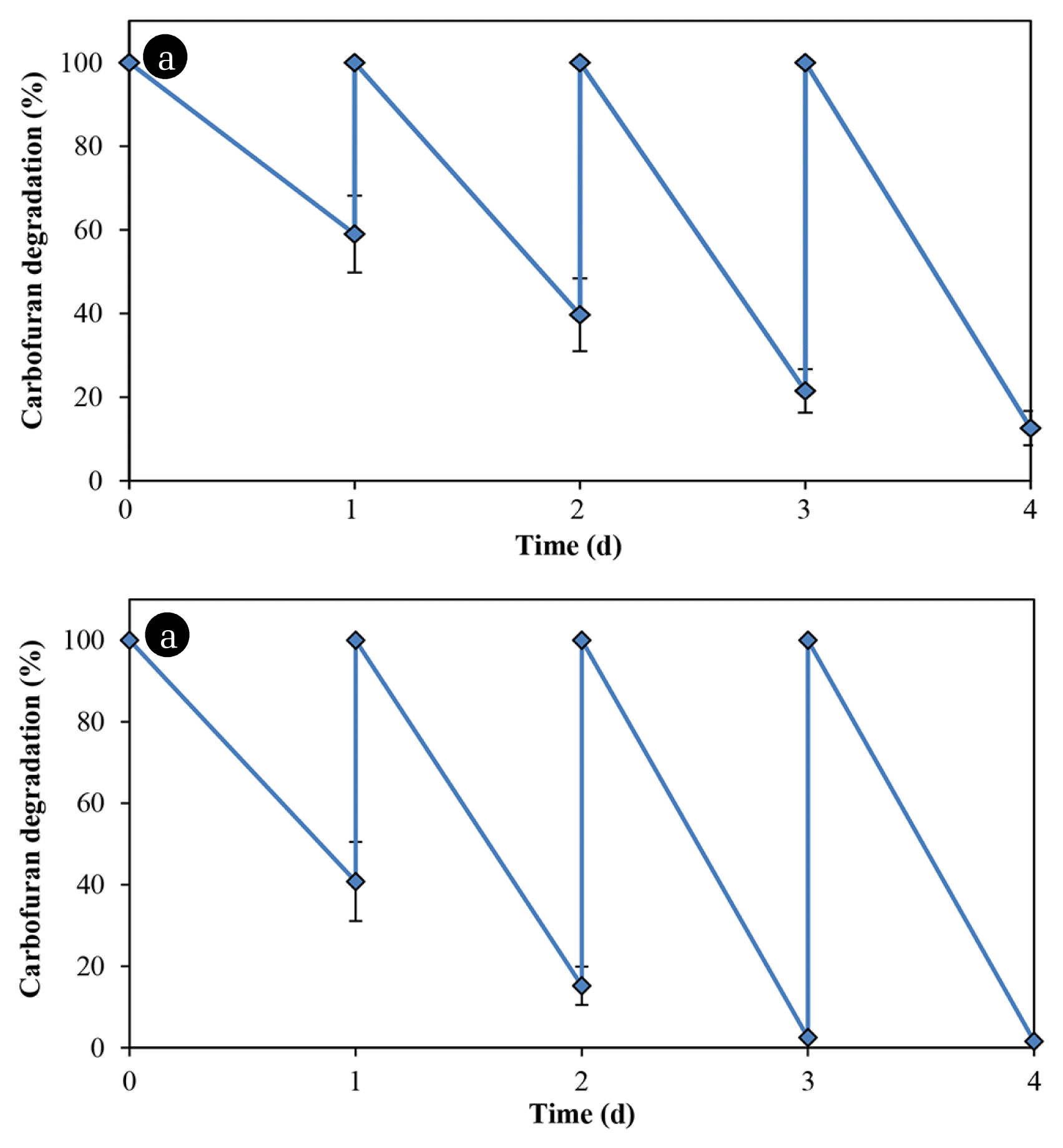
A comparison was made of the carbofuran degradation by bacteria immobilized on glass slides and rice straw. Table 1 lists the results of the analysis of straw components. Although the amount of substrate was absorbed by the rice straw in the first and second cycles, the carbofuran concentration remains unchanged in the subsequent cycles (Fig. 4(a)). Bacillus sp. DT1 could not utilize the compound as a sole nitrogen source but the cells immobilized in the rice straw degraded the carbofuran in media without any co-substrate (Fig. 4(b)). In MM with glucose and ammonium sulfate (0.1% each), the degradation rates of immobilized cells in the rice straw were greater than those of immobilized cells on glass slides (Fig. 5). In this medium, using rice straw as an immobilization material resulting in 97.5% of the substrate was degraded at the 3rd cycle, which was about 20% higher than that of using glass slides.
Numbers of immobilized bacteria by the straw (both the outside surface and the inside) might be greater than the numbers on glass slides because of high porosity of rice straw. The carrier might protect bacteria from chemical toxicity and other environmental stresses. Rice components were also reported to serve as nutrient sources and/or supporting materials for microorganisms in some previous studies. For examples, rice husk was used as a carbon source and a biofilm carrier for denitrification of wastewater [14], was also treated as a lignocellulosic material to increase the growth and activity of fungus Trametes versicolor for carbofuran remediation [6]. Another study showed carbofuran biodegradation by Burkholderia cepacia PCL3 immobilized in a corncob and sugarcane [12]. However, no evidence illustrated that PCL3 used components of the carrier as nutrients to increase the degradation rates.
3.5. Carbofuran Bioremediation in Soils
Table 2 shows that the degradation increased with the augmentation of isolated bacteria, and the degradation rates were found higher in cell immobilization treatments. The results also indicate that carbofuran degradation in sterile samples of previously used carbofuran soil and the first-time-used carbofuran was statistically insignificant. In addition, rice straw was found to boost the chemical dissipation in the non-inoculated and inoculated soil from 7.8% to 10.7% and from 12.5% to 19.8%, respectively. The sterilization of untreated soil resulted in lower degradation by 9.6–11.9%, and corresponding data for treated soil were 16.4–23.6%. The carbofuran degradation in treated soil augmented with Bacillus sp. DT1 and without sterilization was 92.8% on average.
The carbofuran adapted native microbes in the non-sterile soil accounted for higher chemical degradation rates. In this context, one previous report indicated that the increase in soil degradation of microbial carbofuran was caused by repeated application of the pesticide [26]. The carbofuran loss in the soils without bacteria inoculated was probably due to the degradation by indigenous microorganisms and chemical adsorption by soil components and rice straw. A previous study showed that carbofuran removal by coconut fiber, garden compost and pesticide pre-exposed soil was statistically significant from week 36 onwards [17]. In this study, the use of Bacillus sp. DT1 immobilized in rice straw degraded 92.8% of carbofuran in soil within 30 d.
However, biodegradation rates in both untreated and treated soils were comparably slower than those in liquid media. The carbofuran absorption in soils and soil-carbofuran bonds hindered bacterial degradation and the physicochemical properties of the soils influenced biodegradation processes. Although several reports have addressed carbofuran biodegradation in soils [7, 8, 11], only one of them indicated that using immobilized bacteria increased the biodegradation rate [11].
3.6. Metabolites of Carbofuran Degradation
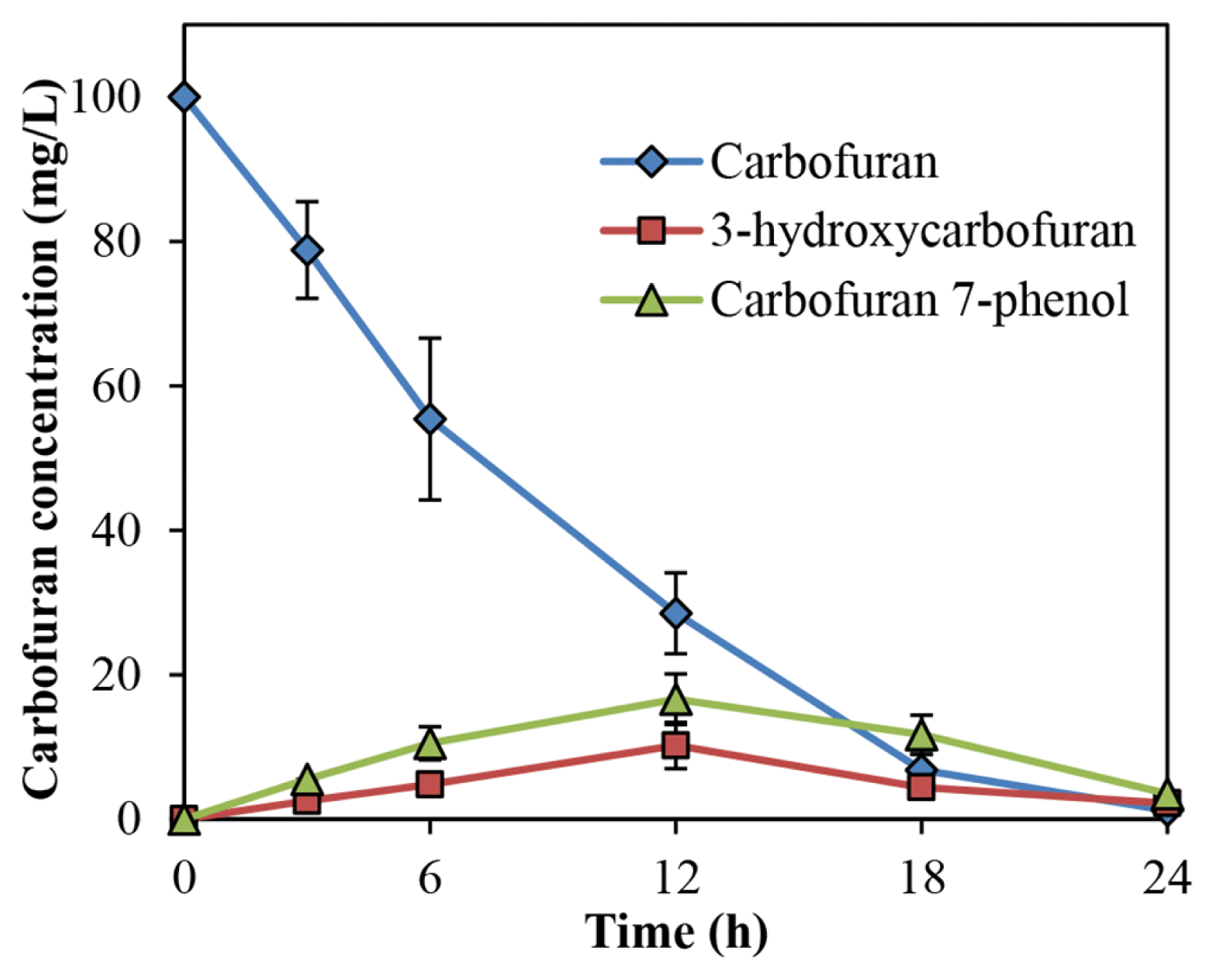
During the carbofuran degradation by Bacillus sp. DT1 in liquid media without straw, several transient metabolites have also been detected. One intermediate (m/z 137, 147 180) (Fig. S1(a)) was proposed to be 3-hydroxycarbofuran, and another product was identified as carbofuran 7-phenol (m/z 131, 149, 164) (Fig. S1(b)). Similarly, m/z 182, 164, 149 (Fig. S1(c)) was identified as 2-hydroxy-3-(3-methypropan-2-ol) phenol [11] during the degradation. The result shows the occurrence of hydrolysis of the furanyl ring. The concentrations of these metabolites were highest at 12 h and gradually went down in the remaining time as presented in Fig. 6. Moreover, Bacillus sp. DT1 degraded as 65.4 ± 7.3% of 3-hydroxycarbofuran with a concentration of 100 mg/L.
For degradation pathways for carbofuran, most bacteria show partial transformation involving hydrolysis without further degradation of the resulting phenolic intermediate [27]. In this study, 3-hydroxycarbofuran, carbofuran 7-phenol and 2-hydroxy-3-(3-methypropan-2-ol) phenol were produced as metabolites of bacterial carbofuran transformation. The appearance of 2-hydroxy-3-(3-methypropan-2-ol) phenol as an intermediate in carbofuran was reported in a previous report [10]. The appearance of 2-hydroxy-3-(3-methypropan-2-ol) phenol indicated the occurrence of hydrolysis of the furanyl ring. It is known that carbofuran 7-phenol and 3-hydroxycarbofuran are metabolites of carbofuran biodegradation [7, 10, 13, 28]. In another report, carbofuran was catabolized by oxidation or hydrolytic pathway producing carbofuran 7-phenol, methylamine, 4-hydroxycarbofuran, 3-ketocarbofuran are produced [11].
4. Conclusions
The present study indicates that Bacillus sp. DT1 is more effective to degrade carbofuran in soils achieved by immobilization in rice straw compared to that by free cells. During the biodegradation, 3-hydroxycarbofuran, carbofuran 7-phenol and 2-hydroxy-3-(3-methypropan-2-ol) phenol were produced as intermediates, which showed the occurrence of the furanyl ring hydrolysis. This work provides a microbial immobilization method which can be productively deployed for treating and safe eliminating toxic pollutants from environments.